For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VMPKEFRFGVSVRNAESQAQVAATARRAEDLGYNVIVVPDHLGAPAPFPTLTAIAAATDTIRVGTCVLNA CFYKPALLARDVTALRDLSDGRFEVGLGAGYVREEFEAAELPFPSARERIDHLRHTTEYLREHVSDVPIM VAGNGDRLLTVAAQTADIVGLTGGDGVPDGDHDPLAERISFLRKAADGRDLELNITFTAIPAAGSEMPDL RLTRRFLGHLTDEQLLRHPGVLAGSASDMADRLRQLRDTYGVSYFIVQIPHIETFAKVIAELR
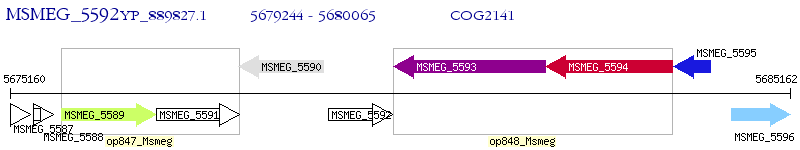
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5592 | - | - | 100% (273) | hypothetical protein MSMEG_5592 |
| M. smegmatis MC2 155 | MSMEG_3977 | - | 2e-45 | 36.01% (286) | hypothetical protein MSMEG_3977 |
| M. smegmatis MC2 155 | MSMEG_2516 | - | 2e-25 | 30.79% (302) | putative oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2917 | - | 8e-33 | 31.95% (313) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_1815 | - | 9e-88 | 59.19% (272) | luciferase family protein |
| M. tuberculosis H37Rv | Rv2893 | - | 9e-33 | 31.95% (313) | oxidoreductase |
| M. leprae Br4923 | MLBr_00269 | - | 4e-05 | 29.57% (115) | putative F420-dependent glucose-6-phosphate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4277 | - | 4e-17 | 38.24% (136) | luciferase-like monooxygenase superfamily protein |
| M. marinum M | MMAR_4587 | - | 8e-83 | 57.72% (272) | oxidoreductase |
| M. avium 104 | MAV_1042 | - | 2e-86 | 59.19% (272) | hypothetical protein MAV_1042 |
| M. thermoresistible (build 8) | TH_2824 | - | 2e-17 | 38.52% (135) | PUTATIVE luciferase family protein |
| M. ulcerans Agy99 | MUL_4447 | - | 4e-39 | 52.23% (157) | hypothetical protein MUL_4447 |
| M. vanbaalenii PYR-1 | Mvan_4930 | - | 8e-96 | 62.73% (271) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2917|M.bovis_AF2122/97 -----------MTVASTAHHTRRLRFG--LAAPLPRAGTQMRAFAQAVEA
Rv2893|M.tuberculosis_H37Rv -----------MTVASTAHHTRRLRFG--LAAPLPRAGTQMRAFAQAVEA
Mflv_1815|M.gilvum_PYR-GCK MPLPPAVQTWTYDHDVAKE----FRFG--VGVTRVSTRAKLEDDARRAEA
Mvan_4930|M.vanbaalenii_PYR-1 ---------------MATE----FRFG--VGVTRARSRAELVDSARRAED
MSMEG_5592|M.smegmatis_MC2_155 --------------MMPKE----FRFG--VSVRNAESQAQVAATARRAED
MMAR_4587|M.marinum_M ---------------MAKD----FRFG--LSVRFIKSRAALQELAKRAED
MUL_4447|M.ulcerans_Agy99 ---------------MLTGRHLYFAYGSNLCVRQMSVRCPDASDPRPAVL
MAV_1042|M.avium_104 ---------------MAKD----FRFG--MSMRFYKSREALLDKAKRAED
MAB_4277|M.abscessus_ATCC_1997 -----------------------MDFRVFVEPQQGATYSDQLRVAQAAEE
TH_2824|M.thermoresistible__bu -----------------------MDFRVFVEPQQGGTYADQLAVARAAED
MLBr_00269|M.leprae_Br4923 --------------------MAELRLGYKASAEQFAPR-ELVELGVAAEA
: .
Mb2917|M.bovis_AF2122/97 AGFDVLAFPDHLVPSVSPFAG-----------------ATAAAMATQRLH
Rv2893|M.tuberculosis_H37Rv AGFDVLAFPDHLVPSVSPFAG-----------------ATAAAMATQRLH
Mflv_1815|M.gilvum_PYR-GCK LGFDVLHVPDHLG-GSAPFP-----------------VMTAVAMATTSLR
Mvan_4930|M.vanbaalenii_PYR-1 LGFDVLHVPDHLG-GPAPFP-----------------VMTTIAMATTTLR
MSMEG_5592|M.smegmatis_MC2_155 LGYNVIVVPDHLG-APAPFP-----------------TLTAIAAATDTIR
MMAR_4587|M.marinum_M LGFDILCVPDHLG-AAAPFP-----------------TLTAAAMVTTKMR
MUL_4447|M.ulcerans_Agy99 SDHDWLINERGVA-TVEPLTGGQVHGVLWQISDRDLATLDSAEGLPVRYR
MAV_1042|M.avium_104 AGFDILCVPDHLG-AAAPFP-----------------TLTAVAMVTTTLR
MAB_4277|M.abscessus_ATCC_1997 LGFSAFFRSDHYLAMGGADGLPG--------PTDAWVTLGAIARETTAIR
TH_2824|M.thermoresistible__bu LGYSAFFRSDHYLAMSG-DGLPG--------PTDSWVTLAGIARETTSIR
MLBr_00269|M.leprae_Br4923 HGMDSATVSDHFQPWRHQGGHAS----------FSLSWMTAVGERTNRIL
. . .
Mb2917|M.bovis_AF2122/97 TGTLVLN-------NDFRHPVDTAR---EAAGVATLAEGRFELGLGAGHR
Rv2893|M.tuberculosis_H37Rv TGTLVLN-------NDFRHPVDTAR---EAAGVATLAEGRFELGLGAGHR
Mflv_1815|M.gilvum_PYR-GCK VGTFVLN-------SAFYRPALLAR---DVAALNDLSDGRVELGLGTGYV
Mvan_4930|M.vanbaalenii_PYR-1 VGTFVIN-------SAFYRPALLAR---DVAALHDLSEGRFELGLGTGYV
MSMEG_5592|M.smegmatis_MC2_155 VGTCVLN-------ACFYKPALLAR---DVTALRDLSDGRFEVGLGAGYV
MMAR_4587|M.marinum_M LSMYVLN-------AAFYKPALLSR---DAEALDLLSEGRLELGLGTGYV
MUL_4447|M.ulcerans_Agy99 RDLLCVRTHDGPTTAWVYIDHRITPGPPRPEYLNRVIDGALHHGLPQRWI
MAV_1042|M.avium_104 LSMYVLN-------SAFYKPALLSR---DMQALDLLSDGRLEIGLGTGYV
MAB_4277|M.abscessus_ATCC_1997 LGTLVTS-------ATFRHPGPLAVT---VAQVDEMSSGRVDFGLGAGWF
TH_2824|M.thermoresistible__bu LGTLVTS-------ATFRYPGPLAIS---VAQVDEMSGGRVEFGLGSGWF
MLBr_00269|M.leprae_Br4923 LGTSVLTP------TFRYNPAVIGQ---AFATMGCLYPNRVFLGVGTGEA
. : : . . *:
Mb2917|M.bovis_AF2122/97 R--SEYD--------------------AAGITFDSG---ATRVARLIESA
Rv2893|M.tuberculosis_H37Rv R--SEYD--------------------AAGITFDSG---ATRVARLIESA
Mflv_1815|M.gilvum_PYR-GCK R--EEFD--------------------AAGIPYP----------------
Mvan_4930|M.vanbaalenii_PYR-1 K--EEFD--------------------AAGIPFP----------------
MSMEG_5592|M.smegmatis_MC2_155 R--EEFE--------------------AAELPFP----------------
MMAR_4587|M.marinum_M R--EEFD--------------------AAELPYP----------------
MUL_4447|M.ulcerans_Agy99 DFLHRWDPVHWLQPVSAAASGAGPQSLSALLSEPG-----------LSEI
MAV_1042|M.avium_104 R--EEFE--------------------AAEIPYP----------------
MAB_4277|M.abscessus_ATCC_1997 S--EEHE--------------------AYAIPFPSLGERFDRLEEQLEIL
TH_2824|M.thermoresistible__bu E--PEHQ--------------------AYAIPFPPLGERFDRLTEQLEII
MLBr_00269|M.leprae_Br4923 LNEVATG---------------------YQGAWPEFKERFARLRESVRLM
.
Mb2917|M.bovis_AF2122/97 HLIRALLDAEPVDFDGQHYRVHAEAGSLVAPPKVRVPLLVGGNG-TEVLR
Rv2893|M.tuberculosis_H37Rv HLIRALLDAEPVDFDGQHYRVHAEAGSLVAPPKVRVPLLVGGNG-TEVLR
Mflv_1815|M.gilvum_PYR-GCK -------------TAGERVDHLKLTAAHIAEHLPDVPIMIAGNG-DRVLR
Mvan_4930|M.vanbaalenii_PYR-1 -------------SARERVDHLKLTIEYMTEHLPDVPIMIAGNG-DRVLR
MSMEG_5592|M.smegmatis_MC2_155 -------------SARERIDHLRHTTEYLREHVSDVPIMVAGNG-DRLLT
MMAR_4587|M.marinum_M -------------SAGARVDYLEHMTAYLKQHHPTVPILIAGNG-NRVMT
MUL_4447|M.ulcerans_Agy99 SQLRSRFGFLAIHGGGARVDYLEHMTAYVKQHHPTVPILIAGNG-NRVMT
MAV_1042|M.avium_104 -------------SARERVDYLEHMTKYLKEHHPSTPLIIAGNG-DRVLT
MAB_4277|M.abscessus_ATCC_1997 TGLWTTPTGDTYSYAGKHYTVTDSPALPKPVQEPHPPIVIGGLGVKRTPA
TH_2824|M.thermoresistible__bu TGLWTTPSGEKYDFTGKHYTVVDSPGLPKPVQDPYPPIIIGGQGPKRTPA
MLBr_00269|M.leprae_Br4923 RELWR---GDRVDFDGDYYQLKG--ASIYDVPEGGVPIYIAAGG-PEVAK
*: :.. * .
Mb2917|M.bovis_AF2122/97 LGGRIADIVGLAGISHNRDATQVRFTHFDADGLADRIAVVRHAAGDRFEA
Rv2893|M.tuberculosis_H37Rv LGGRIADIVGLAGISHNRDATQVRFTHFDADGLADRIAVVRHAAGDRFEA
Mflv_1815|M.gilvum_PYR-GCK TAARFADIVGFTGGDR--------AATGDEDPLGDRIGFVRAAAGERFAA
Mvan_4930|M.vanbaalenii_PYR-1 IAARSADIIGFTGGDR--------AATADEDPLADRIAFVRDAAGERFGD
MSMEG_5592|M.smegmatis_MC2_155 VAAQTADIVGLTGGDG--------VPDGDHDPLAERISFLRKAADGR--D
MMAR_4587|M.marinum_M LAAQQADIVGLTGSN----------VHGADDPLAERVDFVRRAAGDRFDA
MUL_4447|M.ulcerans_Agy99 LAAQQANIVGLTGSN----------VHGADDPLAERVDFVRRAAGDRFDA
MAV_1042|M.avium_104 IAARNADIIGLTGAK----------VRDVEDPFAERVEFVRNAAGDRFDS
MAB_4277|M.abscessus_ATCC_1997 LAARFATEFNLPFQP-----------VDVITDQYARVAHAVEAAGRPADS
TH_2824|M.thermoresistible__bu LAARFANEFNIPFVR-----------LDALKAQFERVAAAVAEAGREPDS
MLBr_00269|M.leprae_Br4923 YAGRAGEGFVCTSGKGEE-----LYTEKLIPAVLEGAAVAGRDADDIDKM
..: . . . *.
Mb2917|M.bovis_AF2122/97 IELNALIQ----------------AVVCTNDRNAAAAELAATLGGITPEQ
Rv2893|M.tuberculosis_H37Rv IELNALIQ----------------AVVCTNDRNAAAAELAATLGGITPEQ
Mflv_1815|M.gilvum_PYR-GCK LELNIAIT----------------AIPLDSSGQPDLTIPRISLPGLSDDE
Mvan_4930|M.vanbaalenii_PYR-1 LELNLAVT----------------AMPVDDSGRPDLTIPRISLPGLSDEE
MSMEG_5592|M.smegmatis_MC2_155 LELNITFT----------------AIPAAGSEMPDLRLTRRFLGHLTDEQ
MMAR_4587|M.marinum_M LELNLAIT----------------ALPNEGETAPDLAMTRRYAPELSDEQ
MUL_4447|M.ulcerans_Agy99 LELNLAIT----------------ALPNEGETAPDLAMTRRYAPELSDEQ
MAV_1042|M.avium_104 LELNLAIT----------------AMPRVGETEPDLKLTRSYSPELTDEQ
MAB_4277|M.abscessus_ATCC_1997 MTYSAAFA----------------LCIGDT----DADIARRAANIGREVE
TH_2824|M.thermoresistible__bu MIYSAAFV----------------LCAGRD----DAEIARRAAAINREVD
MLBr_00269|M.leprae_Br4923 IEIKMSYDPDPEQALSNIRFWAPLSLAAEQKHSIDDPIEMEKVADALPIE
: . :
Mb2917|M.bovis_AF2122/97 VLESPFLLLGTHEQMAEALAARQRRFGVSYWTVFDEWAGRASAMRDIA-E
Rv2893|M.tuberculosis_H37Rv VLESPFLLLGTHEQMAEALAARQRRFGVSYWTVFDEWAGRASAMRDIA-E
Mflv_1815|M.gilvum_PYR-GCK LLRHPSVLSGSTTEIADRIHGYRDVYGISYVIVQMR------HAEAFG-A
Mvan_4930|M.vanbaalenii_PYR-1 LLRHPGVLSGSVDEIADRIRGYRDVYGISYLIVQMR------HAEAFG-K
MSMEG_5592|M.smegmatis_MC2_155 LLRHPGVLAGSASDMADRLRQLRDTYGVSYFIVQIP------HIETFA-K
MMAR_4587|M.marinum_M ILALHSVLGGSPREIADTLAGYRDIYGVTSFTVQDN------HLDNFA-K
MUL_4447|M.ulcerans_Agy99 ILALHSVLGGSPREIADTLAGYRDIYGVTSFTVQDN------HLDNFA-K
MAV_1042|M.avium_104 ILSMPSVLSGSPRDIADTLSAYRDKYGVSSFTVQDN------HIDNFA-K
MAB_4277|M.abscessus_ATCC_1997 EITENSPLAGTPDAVAEKLSIYVD-AGVQRVYVQLLDIRDLDHLEFFAST
TH_2824|M.thermoresistible__bu ELRSNSPVVGTPAEIVDRLGPFIE-AGVQRVYLQLLDMSDLDHLELIATQ
MLBr_00269|M.leprae_Br4923 QVAKRWIVVSDPDEAVARVGQYVT-WGLNHLVFHAPGHNQRRFLELFEKD
: : . . : *: . :
Mb2917|M.bovis_AF2122/97 VIALLRYG-
Rv2893|M.tuberculosis_H37Rv VIALLRYG-
Mflv_1815|M.gilvum_PYR-GCK VIDLLR---
Mvan_4930|M.vanbaalenii_PYR-1 VIELLR---
MSMEG_5592|M.smegmatis_MC2_155 VIAELR---
MMAR_4587|M.marinum_M VIAELR---
MUL_4447|M.ulcerans_Agy99 VIAELR---
MAV_1042|M.avium_104 VIAELR---
MAB_4277|M.abscessus_ATCC_1997 VIPQFS---
TH_2824|M.thermoresistible__bu VVPQLR---
MLBr_00269|M.leprae_Br4923 LAPRLRRLG
: :