For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KFGISTFVNDDSIDPVSLARAIEERGFSSLVIAEHTHIPASRETPYPGGDELPGVYYRTLDPFVTLAAAA AVTSKIELFTGIALLIQRDPITTAKEAASIDLISGGRFVFGVGAGWNIEELRDHGTDPKTRGALLDERIE AIKALWTDEPAEYHGKYVDFPPSYSRPKPVRQPHPPIYIGGNSDATVKRVVRHDAGWISNPLPVHQLTQR VDQLRSAAGHDVPLAMFGTRNKPDYWRAAQDLGFGPAGAAAAHLPAGGVPAAARRIRCPGGSIPPVEVPG GPSFLREPLARVSCASRRKIAPRFAAQCDFASARGGRLRVGSRPR*
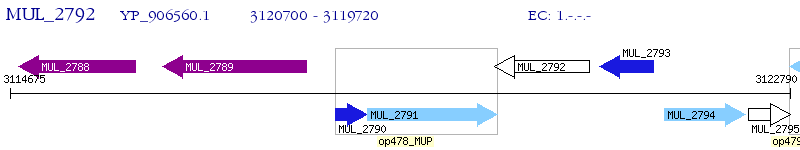
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_2792 | - | - | 100% (326) | hypothetical protein MUL_2792 |
| M. ulcerans Agy99 | MUL_3458 | - | 5e-68 | 53.74% (227) | oxidoreductase |
| M. ulcerans Agy99 | MUL_1043 | - | 2e-30 | 35.83% (187) | hypothetical protein MUL_1043 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3106c | - | 8e-71 | 55.07% (227) | hypothetical protein Mb3106c |
| M. gilvum PYR-GCK | Mflv_1017 | - | 1e-115 | 78.46% (246) | luciferase family protein |
| M. tuberculosis H37Rv | Rv3079c | - | 8e-71 | 55.07% (227) | hypothetical protein Rv3079c |
| M. leprae Br4923 | MLBr_00131 | - | 2e-05 | 26.09% (115) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_1059c | - | 8e-48 | 42.56% (242) | hypothetical protein MAB_1059c |
| M. marinum M | MMAR_3559 | - | 1e-141 | 98.37% (246) | hypothetical protein MMAR_3559 |
| M. avium 104 | MAV_1077 | - | 2e-47 | 41.67% (240) | hypothetical protein MAV_1077 |
| M. smegmatis MC2 155 | MSMEG_1996 | - | 1e-114 | 76.95% (256) | N5,N10- methylenetetrahydromethanopterin reductase-related |
| M. thermoresistible (build 8) | TH_2750 | - | 1e-112 | 76.21% (248) | PUTATIVE luciferase family protein |
| M. vanbaalenii PYR-1 | Mvan_5819 | - | 1e-116 | 80.08% (246) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3106c|M.bovis_AF2122/97 -----------MQFGVLTFVTDEGIGPAELGAALEHR----GFESLFLAE
Rv3079c|M.tuberculosis_H37Rv -----------MQFGVLTFVTDEGIGPAELGAALEHR----GFESLFLAE
MUL_2792|M.ulcerans_Agy99 -----------MKFGISTFVNDDSIDPVSLARAIEER----GFSSLVIAE
MMAR_3559|M.marinum_M -----------MKFGISTFVNDDSIDPVSLARAIEER----GFSSLVIAE
MSMEG_1996|M.smegmatis_MC2_155 MHTVINSQGTGMKIGISTFVNDDTIDAVTLARAIEER----GFHSLVIAE
Mflv_1017|M.gilvum_PYR-GCK -----------MKFGISTFPNEDTIDPVSLARAVEER----GFAALAVAE
Mvan_5819|M.vanbaalenii_PYR-1 -----------MKFGIATFVNDTTIDPVALARAVEDR----GFGSLAVAE
TH_2750|M.thermoresistible__bu -----------MRFGISTFVNDDTIDPVSLATAIEER----GFDALTVAE
MAB_1059c|M.abscessus_ATCC_199 -----------MDYGLVLFTSDRGISPAIAAKAAEDG----GFSTFYVPE
MAV_1077|M.avium_104 ----------------MLFTSDRGISPAAAAKLADDH----GFATFYVPE
MLBr_00131|M.leprae_Br4923 --------MAGFRFGFVDALVHTLFPPSLPARASIVSGAVLGADSYWVGD
. : . . * : : :
Mb3106c|M.bovis_AF2122/97 HTHIPVNTQSPYP---GGGPIP-EKYYRTLDPFVALA--AAAATTQSLVL
Rv3079c|M.tuberculosis_H37Rv HTHIPVNTQSPYP---GGGPIP-EKYYRTLDPFVALA--AAAATTQSLVL
MUL_2792|M.ulcerans_Agy99 HTHIPASRETPYP---GGDELP-GVYYRTLDPFVTLA--AAAAVTSKIEL
MMAR_3559|M.marinum_M HTHIPASRETPYP---GGGELP-GVYYRTLDPFVTLA--AAAAVTSKIEL
MSMEG_1996|M.smegmatis_MC2_155 HSHIPASRESAYP---GGDELP-QKYYRTLDPFVTLA--AAAAVTTKIEL
Mflv_1017|M.gilvum_PYR-GCK HTHIPASRESAYP---LGGELP-KIYYRTLDPFVTLA--AAAAVTSEIEL
Mvan_5819|M.vanbaalenii_PYR-1 HTHIPASRESAYP---LGGELP-SIYYRTLDPFVTLA--AAAAVTTSIEL
TH_2750|M.thermoresistible__bu HTHIPASRESPYP---EGGELP-SVYYRTLDPFVTLA--AAAAVTSRIQL
MAB_1059c|M.abscessus_ATCC_199 HTHIPVKREAAHPG-TGGAELPDDRYMRTLDPWVSLG--AACAVTSKIRL
MAV_1077|M.avium_104 HTHIPIKREAAHPT-TGDASLPDDRYMRTLDPWVSLG--AACAVTSRVRL
MLBr_00131|M.leprae_Br4923 HLNALVPRSVATPKYLGVAAKVVPKIDANYEPWTMLGNLAAGNRLNGLRL
* : . . * . :*:. *. ** : *
Mb3106c|M.bovis_AF2122/97 GTGIALIPERDPIVTAKEVASLDLVSQGRFRFGVGVGWLREEVANHGVDP
Rv3079c|M.tuberculosis_H37Rv GTGIALIPERDPIVTAKEVASLDLVSQGRFRFGVGVGWLREEVANHGVDP
MUL_2792|M.ulcerans_Agy99 FTGIALLIQRDPITTAKEAASIDLISGGRFVFGVGAGWNIEELRDHGTDP
MMAR_3559|M.marinum_M FTGIALLIQRDPITTAKEAASIDLISGGRFVFGVGAGWNIEELRDHGTDP
MSMEG_1996|M.smegmatis_MC2_155 FTGVALLIQRDVIHTAKEAASVDLISNGRFVFGVGAGWNLEEMRNHGTDP
Mflv_1017|M.gilvum_PYR-GCK ITGIALLIQRDPIITAKEAASIDVISGGRFVFGVGAGWNIEELRHHGTDP
Mvan_5819|M.vanbaalenii_PYR-1 VTGIALLIQRDPIITAKEAASIDLISNGRFVFGVGAGWNLEELRHHGTDP
TH_2750|M.thermoresistible__bu ITGIALLIQRDPIITAKEAASIDLISDGRFVFGVGAGWNLEEMRHHGTDP
MAB_1059c|M.abscessus_ATCC_199 ATAVAIPVEHDPITLAKSIATLDHMSGGRVTVGVGYGWNTDELADHHVPA
MAV_1077|M.avium_104 STAVALPVEHDPITLAKSIATLDHLSGGRVSLGVGFGWNTDELADHGVPA
MLBr_00131|M.leprae_Br4923 GVCVTDAGRRNPAVTAQAAATLHLLTRGKAMLGIGVGE-REGNEPYGVEW
. :: .:: *: *::. :: *: .*:* * : : .
Mb3106c|M.bovis_AF2122/97 AVRGRVIDERLRAIIEIWTQ--EQAEFHGTYVDFDPIYCWPKPVTKPYPP
Rv3079c|M.tuberculosis_H37Rv AVRGRVIDERLRAIIEIWTQ--EQAEFHGTYVDFDPIYCWPKPVTKPYPP
MUL_2792|M.ulcerans_Agy99 KTRGALLDERIEAIKALWTD--EPAEYHGKYVDFPPSYSRPKPVRQPHPP
MMAR_3559|M.marinum_M KTRGALLDERIEAIKALWTD--EPAEYHGKYVDFPPSYSRPKPVRQPHPP
MSMEG_1996|M.smegmatis_MC2_155 KTRGRLLDEQIEAIKALWTQ--EPAEYHGKYVDIESSYMRPKPVQKPHPP
Mflv_1017|M.gilvum_PYR-GCK KTRGALLDERIEAIKVLWTQ--EPAEYHGRFVDFDASYQRPKPVRKPHPP
Mvan_5819|M.vanbaalenii_PYR-1 KTRGALLDERIEAIKALWTA--EPAEYHGKYVDFDASYLRPKPVQKPHPP
TH_2750|M.thermoresistible__bu RTRGALLDERIEAVKALWTD--EPAEYHGRFVDFGPSYLRPKPVQKPHPP
MAB_1059c|M.abscessus_ATCC_199 GRRRTMLREYIEAMRRLWEE--EEASYSGEFVNFGPSWAWPKTVQKHVPV
MAV_1077|M.avium_104 GRRRTMLREYLEAMRALWTQ--EEAAYDGEFVKFGPSWAWPKPVQPHIPV
MLBr_00131|M.leprae_Br4923 TKPVARFQEALATIRALWDSNGELVSRESQFFPLHNALFDLPPYRGKWPE
: * : :: :* * . . :. : . *
Mb3106c|M.bovis_AF2122/97 LYVGGGP-ANFPRIARLNAGWIAIS--PSPQR--LSGPLQRLRAMAGGD-
Rv3079c|M.tuberculosis_H37Rv LYVGGGP-ANFPRIARLNAGWIAIS--PSPQR--LSGPLQRLRAMAGGD-
MUL_2792|M.ulcerans_Agy99 IYIGGNSDATVKRVVRHDAGWISNP--LPVHQ--LTQRVDQLRSAAGHD-
MMAR_3559|M.marinum_M IYIGGNSDATVKRVVRHDAGWISNP--LPVRQ--LTQRVDQLRGAAGHD-
MSMEG_1996|M.smegmatis_MC2_155 IYIGGASEATVKRVIRHDAGWISNP--LPVDF--LSKFVTQMRDGLGHE-
Mflv_1017|M.gilvum_PYR-GCK ILIGGDSDATVRRIIRHGAGWIANP--LPIDH--LSRRIEQIREGAGGD-
Mvan_5819|M.vanbaalenii_PYR-1 ILIGGDSDAAVKRIIRHEAGWISNP--LPVDT--LRRRVDQIRAGARHE-
TH_2750|M.thermoresistible__bu IVVGGNSDATVRRIIRHGAGWMSNP--HPPEM--LKRRVQQIRDGAGHD-
MAB_1059c|M.abscessus_ATCC_199 IVGAGGTEKNFTWIAKNADGWMSTPRDEDIAG--KIVALKEAWKAAGRDG
MAV_1077|M.avium_104 LVGAAGNEKNFKWIARSADGWITTPRDFDIDE--PVKLLQDTWAAAGRDG
MLBr_00131|M.leprae_Br4923 IWVAAHGPRMLQATGRYADAWIPIVLVRPTDYSCALEVVRTAASDAGRDP
: .. . : .*:. : :
Mb3106c|M.bovis_AF2122/97 VPVTVCQWG-----EAAAKDLEGYRHLGVE---RVLLELPT---------
Rv3079c|M.tuberculosis_H37Rv VPVTVCQWG-----EAAAKDLEGYRHLGVE---RVLLELPT---------
MUL_2792|M.ulcerans_Agy99 VPLAMFGT------RNKPDYWRAAQDLGFGPAGAAAAHLPAGGVPAAARR
MMAR_3559|M.marinum_M VPLAMFGT------PNKPDYWRAAQDLGFG---QLALLLPT---------
MSMEG_1996|M.smegmatis_MC2_155 VPLAQFGT------PADPEYWGAVEELGFG---QIALLLPT---------
Mflv_1017|M.gilvum_PYR-GCK VPLCQFGT------PVKPEYWQAAEELGFG---QLNLLLPT---------
Mvan_5819|M.vanbaalenii_PYR-1 VPLSMFGT------PVKPEYWQAAEDLGFG---QLNLLLPT---------
TH_2750|M.thermoresistible__bu VLLTTFGT------PVDPGYWEVLEELGYQ---QANLLLPT---------
MAB_1059c|M.abscessus_ATCC_199 DPLIYVLAG-----KPDPEQLAAWRELGVT---EAIFGMPD---------
MAV_1077|M.avium_104 APQIVALDF-----KPDAEKLARWAELGVT---EVLFGLPD---------
MLBr_00131|M.leprae_Br4923 MSITPAAVRGIITGRTRDDVDEALDSVLVR---MIALGVPGEAWARHGVE
: :*
Mb3106c|M.bovis_AF2122/97 --EPRD---------------PTLRYLDKLQAELARLA------------
Rv3079c|M.tuberculosis_H37Rv --EPRD---------------PTLRYLDKLQAELARLA------------
MUL_2792|M.ulcerans_Agy99 IRCPGGSI-------PPVEVPGGPSFLREPLARVSCASRRKIAPRFAAQC
MMAR_3559|M.marinum_M --CPRD---------------ESLRLLDEYAALVAQYHR-----------
MSMEG_1996|M.smegmatis_MC2_155 --LPTD---------------ESLRMLDDYATAVEKYRG-----------
Mflv_1017|M.gilvum_PYR-GCK --KSRD---------------VSLRLLDDYAEQVARYIG-----------
Mvan_5819|M.vanbaalenii_PYR-1 --KPRD---------------ESLRLLDGYAEQVARY-------------
TH_2750|M.thermoresistible__bu --RPRD---------------ESLRMLDQFAERVAGYRG-----------
MAB_1059c|M.abscessus_ATCC_199 --RSEE---------------EVRAYIDRLSGKLAAFA------------
MAV_1077|M.avium_104 --KSED---------------EVAAYVERLAGKLAALV------------
MLBr_00131|M.leprae_Br4923 HPMGADFAGVQDIIPQTIDEETVVSYAAKVPAALMKEVLFSGTPEEVIDQ
:
Mb3106c|M.bovis_AF2122/97 --------------------------------------------
Rv3079c|M.tuberculosis_H37Rv --------------------------------------------
MUL_2792|M.ulcerans_Agy99 DFASARGGRLRVGSRPR---------------------------
MMAR_3559|M.marinum_M --------------------------------------------
MSMEG_1996|M.smegmatis_MC2_155 --------------------------------------------
Mflv_1017|M.gilvum_PYR-GCK --------------------------------------------
Mvan_5819|M.vanbaalenii_PYR-1 --------------------------------------------
TH_2750|M.thermoresistible__bu --------------------------------------------
MAB_1059c|M.abscessus_ATCC_199 --------------------------------------------
MAV_1077|M.avium_104 --------------------------------------------
MLBr_00131|M.leprae_Br4923 VAEWRDHGLKYLVVINGSLVNSSLRKTVSALLPHARVLRGLKKL