For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RVGVLTFVTDEGIGPAELGAALEERGFESLFLAEHTHIPVNNQSPYPPGGPIPRKYYRTLDPFVALTAAA VATESLVLGTGIALIPERDPIVTAKEVASLDVVSGGRFCFGVGVGWLREEVANHGVDPAVRGRVSDERLG AMIEIWTQDNAEYHGKYVDFDPICCWPKPVTKPHPPLYIGGGPASFGRIARLGAGWIAISPSPQLLAGPL EELRSLAGEEVAVTVCLSDETKVSDLEGYLPLGVERVLLGLPTEPRDQTLRYLDQMESELVQLG*
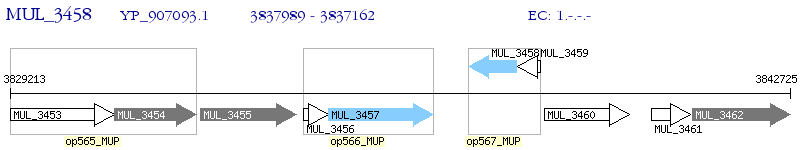
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_3458 | - | - | 100% (275) | oxidoreductase |
| M. ulcerans Agy99 | MUL_2792 | - | 4e-68 | 53.74% (227) | hypothetical protein MUL_2792 |
| M. ulcerans Agy99 | MUL_1043 | - | 3e-32 | 32.42% (293) | hypothetical protein MUL_1043 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3106c | - | 1e-129 | 81.02% (274) | hypothetical protein Mb3106c |
| M. gilvum PYR-GCK | Mflv_1017 | - | 3e-69 | 49.27% (274) | luciferase family protein |
| M. tuberculosis H37Rv | Rv3079c | - | 1e-129 | 81.02% (274) | hypothetical protein Rv3079c |
| M. leprae Br4923 | MLBr_00269 | - | 6e-07 | 26.84% (190) | putative F420-dependent glucose-6-phosphate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1059c | - | 4e-44 | 35.71% (280) | hypothetical protein MAB_1059c |
| M. marinum M | MMAR_1575 | - | 1e-160 | 99.27% (275) | oxidoreductase |
| M. avium 104 | MAV_1077 | - | 2e-46 | 38.41% (276) | hypothetical protein MAV_1077 |
| M. smegmatis MC2 155 | MSMEG_1996 | - | 6e-67 | 49.07% (269) | N5,N10- methylenetetrahydromethanopterin reductase-related |
| M. thermoresistible (build 8) | TH_2750 | - | 3e-74 | 50.94% (267) | PUTATIVE luciferase family protein |
| M. vanbaalenii PYR-1 | Mvan_5819 | - | 2e-69 | 50.75% (266) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_3458|M.ulcerans_Agy99 -----------MRVGVLTFVTDEGIGP---AELGAALEERGFESLFLAEH
MMAR_1575|M.marinum_M -----------MRVGVLTFVTDEGIGP---AELGAALEERGFESLFLAEH
Mb3106c|M.bovis_AF2122/97 -----------MQFGVLTFVTDEGIGP---AELGAALEHRGFESLFLAEH
Rv3079c|M.tuberculosis_H37Rv -----------MQFGVLTFVTDEGIGP---AELGAALEHRGFESLFLAEH
Mflv_1017|M.gilvum_PYR-GCK -----------MKFGISTFPNEDTIDP---VSLARAVEERGFAALAVAEH
Mvan_5819|M.vanbaalenii_PYR-1 -----------MKFGIATFVNDTTIDP---VALARAVEDRGFGSLAVAEH
MSMEG_1996|M.smegmatis_MC2_155 MHTVINSQGTGMKIGISTFVNDDTIDA---VTLARAIEERGFHSLVIAEH
TH_2750|M.thermoresistible__bu -----------MRFGISTFVNDDTIDP---VSLATAIEERGFDALTVAEH
MAB_1059c|M.abscessus_ATCC_199 -----------MDYGLVLFTSDRGISP---AIAAKAAEDGGFSTFYVPEH
MAV_1077|M.avium_104 ----------------MLFTSDRGISP---AAAAKLADDHGFATFYVPEH
MLBr_00269|M.leprae_Br4923 ----------MAELRLGYKASAEQFAPRELVELGVAAEAHGMDSATVSDH
. : . . . : *: : :.:*
MUL_3458|M.ulcerans_Agy99 THIPVNNQSPYP---PGGPIP-RKYYRTLDPFVALTAAAVATESLVLGTG
MMAR_1575|M.marinum_M THIPVNNQSPYP---PGGPIP-RKYYRTLDPFVALTAAAVATESLVLGTG
Mb3106c|M.bovis_AF2122/97 THIPVNTQSPYP---GGGPIP-EKYYRTLDPFVALAAAAATTQSLVLGTG
Rv3079c|M.tuberculosis_H37Rv THIPVNTQSPYP---GGGPIP-EKYYRTLDPFVALAAAAATTQSLVLGTG
Mflv_1017|M.gilvum_PYR-GCK THIPASRESAYP---LGGELP-KIYYRTLDPFVTLAAAAAVTSEIELITG
Mvan_5819|M.vanbaalenii_PYR-1 THIPASRESAYP---LGGELP-SIYYRTLDPFVTLAAAAAVTTSIELVTG
MSMEG_1996|M.smegmatis_MC2_155 SHIPASRESAYP---GGDELP-QKYYRTLDPFVTLAAAAAVTTKIELFTG
TH_2750|M.thermoresistible__bu THIPASRESPYP---EGGELP-SVYYRTLDPFVTLAAAAAVTSRIQLITG
MAB_1059c|M.abscessus_ATCC_199 THIPVKREAAHPG-TGGAELPDDRYMRTLDPWVSLGAACAVTSKIRLATA
MAV_1077|M.avium_104 THIPIKREAAHPT-TGDASLPDDRYMRTLDPWVSLGAACAVTSRVRLSTA
MLBr_00269|M.leprae_Br4923 FQPWRHQGGHASFS-----------------LSWMTAVGERTNRILLGTS
: . . : *. * : * *.
MUL_3458|M.ulcerans_Agy99 IALIPER-DPIVTAKEVASLDVVSGGRFCFGVGVGWLREEVAN--HGVDP
MMAR_1575|M.marinum_M IALIPER-DPIVTAKEVASLDVVSGGRFCFGVGVGWLREEVAN--HGVDP
Mb3106c|M.bovis_AF2122/97 IALIPER-DPIVTAKEVASLDLVSQGRFRFGVGVGWLREEVAN--HGVDP
Rv3079c|M.tuberculosis_H37Rv IALIPER-DPIVTAKEVASLDLVSQGRFRFGVGVGWLREEVAN--HGVDP
Mflv_1017|M.gilvum_PYR-GCK IALLIQR-DPIITAKEAASIDVISGGRFVFGVGAGWNIEELRH--HGTDP
Mvan_5819|M.vanbaalenii_PYR-1 IALLIQR-DPIITAKEAASIDLISNGRFVFGVGAGWNLEELRH--HGTDP
MSMEG_1996|M.smegmatis_MC2_155 VALLIQR-DVIHTAKEAASVDLISNGRFVFGVGAGWNLEEMRN--HGTDP
TH_2750|M.thermoresistible__bu IALLIQR-DPIITAKEAASIDLISDGRFVFGVGAGWNLEEMRH--HGTDP
MAB_1059c|M.abscessus_ATCC_199 VAIPVEH-DPITLAKSIATLDHMSGGRVTVGVGYGWNTDELAD--HHVPA
MAV_1077|M.avium_104 VALPVEH-DPITLAKSIATLDHLSGGRVSLGVGFGWNTDELAD--HGVPA
MLBr_00269|M.leprae_Br4923 VLTPTFRYNPAVIGQAFATMGCLYPNRVFLGVGTGEALNEVATGYQGAWP
: : : .: *::. : .*. .*** * :*: : . .
MUL_3458|M.ulcerans_Agy99 AVRG--RVSDERLGAMIEIWTQDNAEYHGKYVDFDPICCWPKPVTKPHPP
MMAR_1575|M.marinum_M AVRG--RVTDERLGAMIEIWTQDNAEYHGKYVDFDPICCWPKPVTKPHPP
Mb3106c|M.bovis_AF2122/97 AVRG--RVIDERLRAIIEIWTQEQAEFHGTYVDFDPIYCWPKPVTKPYPP
Rv3079c|M.tuberculosis_H37Rv AVRG--RVIDERLRAIIEIWTQEQAEFHGTYVDFDPIYCWPKPVTKPYPP
Mflv_1017|M.gilvum_PYR-GCK KTRG--ALLDERIEAIKVLWTQEPAEYHGRFVDFDASYQRPKPVRKPHPP
Mvan_5819|M.vanbaalenii_PYR-1 KTRG--ALLDERIEAIKALWTAEPAEYHGKYVDFDASYLRPKPVQKPHPP
MSMEG_1996|M.smegmatis_MC2_155 KTRG--RLLDEQIEAIKALWTQEPAEYHGKYVDIESSYMRPKPVQKPHPP
TH_2750|M.thermoresistible__bu RTRG--ALLDERIEAVKALWTDEPAEYHGRFVDFGPSYLRPKPVQKPHPP
MAB_1059c|M.abscessus_ATCC_199 GRRR--TMLREYIEAMRRLWEEEEASYSGEFVNFGPSWAWPKTVQKHVPV
MAV_1077|M.avium_104 GRRR--TMLREYLEAMRALWTQEEAAYDGEFVKFGPSWAWPKPVQPHIPV
MLBr_00269|M.leprae_Br4923 EFKERFARLRESVRLMRELWRGDRVDFDGDYYQLKGASIYDVPEGG--VP
: * : : :* : . : * : .: .
MUL_3458|M.ulcerans_Agy99 LYIGGGP-ASFGRIARLGAGWIAISPSPQLLA--GPLEELRSLAGEE---
MMAR_1575|M.marinum_M LYIGGGP-ASFGRIARLGAGWIAISPSPQLLA--GPLEELRSLAGED---
Mb3106c|M.bovis_AF2122/97 LYVGGGP-ANFPRIARLNAGWIAISPSPQRLS--GPLQRLRAMAGGD---
Rv3079c|M.tuberculosis_H37Rv LYVGGGP-ANFPRIARLNAGWIAISPSPQRLS--GPLQRLRAMAGGD---
Mflv_1017|M.gilvum_PYR-GCK ILIGGDSDATVRRIIRHGAGWIANPLPIDHLS--RRIEQIREGAGGD---
Mvan_5819|M.vanbaalenii_PYR-1 ILIGGDSDAAVKRIIRHEAGWISNPLPVDTLR--RRVDQIRAGARHE---
MSMEG_1996|M.smegmatis_MC2_155 IYIGGASEATVKRVIRHDAGWISNPLPVDFLS--KFVTQMRDGLGHE---
TH_2750|M.thermoresistible__bu IVVGGNSDATVRRIIRHGAGWMSNPHPPEMLK--RRVQQIRDGAGHD---
MAB_1059c|M.abscessus_ATCC_199 IVGAGGTEKNFTWIAKNADGWMSTPRDEDIAGKIVALKEAWKAAGRDG--
MAV_1077|M.avium_104 LVGAAGNEKNFKWIARSADGWITTPRDFDIDEPVKLLQDTWAAAGRDG--
MLBr_00269|M.leprae_Br4923 IYIAAGGPEVAKYAGRAGEGFVCTSGKGEELYTEKLIPAVLEGAAVAGRD
: .. : *:: . : :
MUL_3458|M.ulcerans_Agy99 ----------------------------VAVTVCLSDETKVSD-------
MMAR_1575|M.marinum_M ----------------------------VAVTVCLSDETKVSD-------
Mb3106c|M.bovis_AF2122/97 ----------------------------VPVTVCQWGEAAAKD-------
Rv3079c|M.tuberculosis_H37Rv ----------------------------VPVTVCQWGEAAAKD-------
Mflv_1017|M.gilvum_PYR-GCK ----------------------------VPLCQFG-TPVKPEY-------
Mvan_5819|M.vanbaalenii_PYR-1 ----------------------------VPLSMFG-TPVKPEY-------
MSMEG_1996|M.smegmatis_MC2_155 ----------------------------VPLAQFG-TPADPEY-------
TH_2750|M.thermoresistible__bu ----------------------------VLLTTFG-TPVDPGY-------
MAB_1059c|M.abscessus_ATCC_199 ----------------------------DPLIYVLAGKPDPEQ-------
MAV_1077|M.avium_104 ----------------------------APQIVALDFKPDAEK-------
MLBr_00269|M.leprae_Br4923 ADDIDKMIEIKMSYDPDPEQALSNIRFWAPLSLAAEQKHSIDDPIEMEKV
MUL_3458|M.ulcerans_Agy99 -------------------------LEGYLPLGVERVLLGLPTEPRDQTL
MMAR_1575|M.marinum_M -------------------------LEGYLPLGVERVLLGLPTEPRDQTL
Mb3106c|M.bovis_AF2122/97 -------------------------LEGYRHLGVERVLLELPTEPRDPTL
Rv3079c|M.tuberculosis_H37Rv -------------------------LEGYRHLGVERVLLELPTEPRDPTL
Mflv_1017|M.gilvum_PYR-GCK -------------------------WQAAEELGFGQLNLLLPTKSRDVSL
Mvan_5819|M.vanbaalenii_PYR-1 -------------------------WQAAEDLGFGQLNLLLPTKPRDESL
MSMEG_1996|M.smegmatis_MC2_155 -------------------------WGAVEELGFGQIALLLPTLPTDESL
TH_2750|M.thermoresistible__bu -------------------------WEVLEELGYQQANLLLPTRPRDESL
MAB_1059c|M.abscessus_ATCC_199 -------------------------LAAWRELGVTEAIFGMPDRSEEEVR
MAV_1077|M.avium_104 -------------------------LARWAELGVTEVLFGLPDKSEDEVA
MLBr_00269|M.leprae_Br4923 ADALPIEQVAKRWIVVSDPDEAVARVGQYVTWGLNHLVFHAPGHNQRRFL
* . : *
MUL_3458|M.ulcerans_Agy99 RYLDQ-MESELVQLG-
MMAR_1575|M.marinum_M RYLDQ-MESELVQLG-
Mb3106c|M.bovis_AF2122/97 RYLDK-LQAELARLA-
Rv3079c|M.tuberculosis_H37Rv RYLDK-LQAELARLA-
Mflv_1017|M.gilvum_PYR-GCK RLLDD-YAEQVARYIG
Mvan_5819|M.vanbaalenii_PYR-1 RLLDG-YAEQVARY--
MSMEG_1996|M.smegmatis_MC2_155 RMLDD-YATAVEKYRG
TH_2750|M.thermoresistible__bu RMLDQ-FAERVAGYRG
MAB_1059c|M.abscessus_ATCC_199 AYIDR-LSGKLAAFA-
MAV_1077|M.avium_104 AYVER-LAGKLAALV-
MLBr_00269|M.leprae_Br4923 ELFEKDLAPRLRRLG-
.: :