For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MHTVINSQGTGMKIGISTFVNDDTIDAVTLARAIEERGFHSLVIAEHSHIPASRESAYPGGDELPQKYYR TLDPFVTLAAAAAVTTKIELFTGVALLIQRDVIHTAKEAASVDLISNGRFVFGVGAGWNLEEMRNHGTDP KTRGRLLDEQIEAIKALWTQEPAEYHGKYVDIESSYMRPKPVQKPHPPIYIGGASEATVKRVIRHDAGWI SNPLPVDFLSKFVTQMRDGLGHEVPLAQFGTPADPEYWGAVEELGFGQIALLLPTLPTDESLRMLDDYAT AVEKYRG
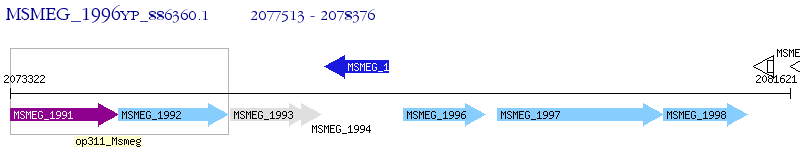
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1996 | - | - | 100% (287) | N5,N10- methylenetetrahydromethanopterin reductase-related protein |
| M. smegmatis MC2 155 | MSMEG_6703 | - | 2e-44 | 40.58% (276) | N5,N10- methylenetetrahydromethanopterin reductase-related |
| M. smegmatis MC2 155 | MSMEG_5520 | - | 3e-41 | 41.90% (210) | hypothetical protein MSMEG_5520 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3106c | - | 8e-71 | 50.94% (267) | hypothetical protein Mb3106c |
| M. gilvum PYR-GCK | Mflv_1017 | - | 1e-121 | 73.91% (276) | luciferase family protein |
| M. tuberculosis H37Rv | Rv3079c | - | 9e-71 | 50.94% (267) | hypothetical protein Rv3079c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4908c | - | 6e-44 | 39.42% (274) | putative luciferase-like oxidoreductase |
| M. marinum M | MMAR_3559 | - | 1e-125 | 78.10% (274) | hypothetical protein MMAR_3559 |
| M. avium 104 | MAV_1077 | - | 2e-42 | 39.54% (263) | hypothetical protein MAV_1077 |
| M. thermoresistible (build 8) | TH_2750 | - | 1e-118 | 72.83% (276) | PUTATIVE luciferase family protein |
| M. ulcerans Agy99 | MUL_2792 | - | 1e-114 | 76.95% (256) | hypothetical protein MUL_2792 |
| M. vanbaalenii PYR-1 | Mvan_5819 | - | 1e-123 | 77.37% (274) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3106c|M.bovis_AF2122/97 -----------MQFGVLTFVTDEGIGPAELGAALEHRGFESLFLAEHTHI
Rv3079c|M.tuberculosis_H37Rv -----------MQFGVLTFVTDEGIGPAELGAALEHRGFESLFLAEHTHI
MMAR_3559|M.marinum_M -----------MKFGISTFVNDDSIDPVSLARAIEERGFSSLVIAEHTHI
MUL_2792|M.ulcerans_Agy99 -----------MKFGISTFVNDDSIDPVSLARAIEERGFSSLVIAEHTHI
MSMEG_1996|M.smegmatis_MC2_155 MHTVINSQGTGMKIGISTFVNDDTIDAVTLARAIEERGFHSLVIAEHSHI
Mflv_1017|M.gilvum_PYR-GCK -----------MKFGISTFPNEDTIDPVSLARAVEERGFAALAVAEHTHI
Mvan_5819|M.vanbaalenii_PYR-1 -----------MKFGIATFVNDTTIDPVALARAVEDRGFGSLAVAEHTHI
TH_2750|M.thermoresistible__bu -----------MRFGISTFVNDDTIDPVSLATAIEERGFDALTVAEHTHI
MAB_4908c|M.abscessus_ATCC_199 ----------MTGFGISTTSADGYVSPDVLARAVEERGFEWLLFDDHSYF
MAV_1077|M.avium_104 ----------------MLFTSDRGISPAAAAKLADDHGFATFYVPEHTHI
: :.. . :.:** : . :*:::
Mb3106c|M.bovis_AF2122/97 PVNTQSPYPG--GGPIP-EKYYRTLDPFVALAAAAATTQSLVLGTGIALI
Rv3079c|M.tuberculosis_H37Rv PVNTQSPYPG--GGPIP-EKYYRTLDPFVALAAAAATTQSLVLGTGIALI
MMAR_3559|M.marinum_M PASRETPYPG--GGELP-GVYYRTLDPFVTLAAAAAVTSKIELFTGIALL
MUL_2792|M.ulcerans_Agy99 PASRETPYPG--GDELP-GVYYRTLDPFVTLAAAAAVTSKIELFTGIALL
MSMEG_1996|M.smegmatis_MC2_155 PASRESAYPG--GDELP-QKYYRTLDPFVTLAAAAAVTTKIELFTGVALL
Mflv_1017|M.gilvum_PYR-GCK PASRESAYPL--GGELP-KIYYRTLDPFVTLAAAAAVTSEIELITGIALL
Mvan_5819|M.vanbaalenii_PYR-1 PASRESAYPL--GGELP-SIYYRTLDPFVTLAAAAAVTTSIELVTGIALL
TH_2750|M.thermoresistible__bu PASRESPYPE--GGELP-SVYYRTLDPFVTLAAAAAVTSRIQLITGIALL
MAB_4908c|M.abscessus_ATCC_199 PVD--TPDSI--DAEFR-ARLPLIRDLPVVLAYAASATERLIVGSGVALL
MAV_1077|M.avium_104 PIKREAAHPTTGDASLPDDRYMRTLDPWVSLGAACAVTSRVRLSTAVALP
* . :. . . : * * *. *.:.* : : :.:**
Mb3106c|M.bovis_AF2122/97 PERDPIVTAKEVASLDLVSQGRFRFGVGVGWLREEVANHGVDPAVRGRVI
Rv3079c|M.tuberculosis_H37Rv PERDPIVTAKEVASLDLVSQGRFRFGVGVGWLREEVANHGVDPAVRGRVI
MMAR_3559|M.marinum_M IQRDPITTAKEAASIDLISGGRFVFGVGAGWNIEELRDHGTDPKTRGALL
MUL_2792|M.ulcerans_Agy99 IQRDPITTAKEAASIDLISGGRFVFGVGAGWNIEELRDHGTDPKTRGALL
MSMEG_1996|M.smegmatis_MC2_155 IQRDVIHTAKEAASVDLISNGRFVFGVGAGWNLEEMRNHGTDPKTRGRLL
Mflv_1017|M.gilvum_PYR-GCK IQRDPIITAKEAASIDVISGGRFVFGVGAGWNIEELRHHGTDPKTRGALL
Mvan_5819|M.vanbaalenii_PYR-1 IQRDPIITAKEAASIDLISNGRFVFGVGAGWNLEELRHHGTDPKTRGALL
TH_2750|M.thermoresistible__bu IQRDPIITAKEAASIDLISDGRFVFGVGAGWNLEEMRHHGTDPRTRGALL
MAB_4908c|M.abscessus_ATCC_199 PQRDVLHTAKAWATVATLSGGRTVLGVGVGWNLGEVRNHGIDPAKRGAVL
MAV_1077|M.avium_104 VEHDPITLAKSIATLDHLSGGRVSLGVGFGWNTDELADHGVPAGRRRTML
::* : ** *:: :* ** :*** ** *: .** . * ::
Mb3106c|M.bovis_AF2122/97 DERLRAIIEIWTQEQAEFHGTYVDFDPIYCWPKPVTKPYPPLYVGGGP-A
Rv3079c|M.tuberculosis_H37Rv DERLRAIIEIWTQEQAEFHGTYVDFDPIYCWPKPVTKPYPPLYVGGGP-A
MMAR_3559|M.marinum_M DERIEAIKALWTDEPAEYHGKYVDFPPSYSRPKPVRQPHPPIYIGGNSDA
MUL_2792|M.ulcerans_Agy99 DERIEAIKALWTDEPAEYHGKYVDFPPSYSRPKPVRQPHPPIYIGGNSDA
MSMEG_1996|M.smegmatis_MC2_155 DEQIEAIKALWTQEPAEYHGKYVDIESSYMRPKPVQKPHPPIYIGGASEA
Mflv_1017|M.gilvum_PYR-GCK DERIEAIKVLWTQEPAEYHGRFVDFDASYQRPKPVRKPHPPILIGGDSDA
Mvan_5819|M.vanbaalenii_PYR-1 DERIEAIKALWTAEPAEYHGKYVDFDASYLRPKPVQKPHPPILIGGDSDA
TH_2750|M.thermoresistible__bu DERIEAVKALWTDEPAEYHGRFVDFGPSYLRPKPVQKPHPPIVVGGNSDA
MAB_4908c|M.abscessus_ATCC_199 DEQLAALHALWTQEQAEFHGEHVDFGPVVARPRPARP--LPVHVGGPSRA
MAV_1077|M.avium_104 REYLEAMRALWTQEEAAYDGEFVKFGPSWAWPKPVQPHIPVLVGAAGNEK
* : *: :** * * :.* .*.: . *:*. : ..
Mb3106c|M.bovis_AF2122/97 NFPRIARLNAGWIAIS--PSPQRLSGPLQRLRAMAGGD-VPVTVCQWGEA
Rv3079c|M.tuberculosis_H37Rv NFPRIARLNAGWIAIS--PSPQRLSGPLQRLRAMAGGD-VPVTVCQWGEA
MMAR_3559|M.marinum_M TVKRVVRHDAGWISNP--LPVRQLTQRVDQLRGAAGHD-VPLAMFGT-PN
MUL_2792|M.ulcerans_Agy99 TVKRVVRHDAGWISNP--LPVHQLTQRVDQLRSAAGHD-VPLAMFGT-RN
MSMEG_1996|M.smegmatis_MC2_155 TVKRVIRHDAGWISNP--LPVDFLSKFVTQMRDGLGHE-VPLAQFGT-PA
Mflv_1017|M.gilvum_PYR-GCK TVRRIIRHGAGWIANP--LPIDHLSRRIEQIREGAGGD-VPLCQFGT-PV
Mvan_5819|M.vanbaalenii_PYR-1 AVKRIIRHEAGWISNP--LPVDTLRRRVDQIRAGARHE-VPLSMFGT-PV
TH_2750|M.thermoresistible__bu TVRRIIRHGAGWMSNP--HPPEMLKRRVQQIRDGAGHD-VLLTTFGT-PV
MAB_4908c|M.abscessus_ATCC_199 AQARALRVGDGWLPYAGTSTPDDVRRACGWFAEQGRAD-LPVTVSGV---
MAV_1077|M.avium_104 NFKWIARSADGWITTPRDFDIDEPVKLLQDTWAAAGRDGAPQIVALDFKP
* **:. . :
Mb3106c|M.bovis_AF2122/97 AAKDLEGYRHLGVE---RVLLELPT-----------EPRD--------PT
Rv3079c|M.tuberculosis_H37Rv AAKDLEGYRHLGVE---RVLLELPT-----------EPRD--------PT
MMAR_3559|M.marinum_M KPDYWRAAQDLGFG---QLALLLPT-----------CPRD--------ES
MUL_2792|M.ulcerans_Agy99 KPDYWRAAQDLGFGPAGAAAAHLPAGGVPAAARRIRCPGGSIPPVEVPGG
MSMEG_1996|M.smegmatis_MC2_155 DPEYWGAVEELGFG---QIALLLPT-----------LPTD--------ES
Mflv_1017|M.gilvum_PYR-GCK KPEYWQAAEELGFG---QLNLLLPT-----------KSRD--------VS
Mvan_5819|M.vanbaalenii_PYR-1 KPEYWQAAEDLGFG---QLNLLLPT-----------KPRD--------ES
TH_2750|M.thermoresistible__bu DPGYWEVLEELGYQ---QANLLLPT-----------RPRD--------ES
MAB_4908c|M.abscessus_ATCC_199 PGDHQLAGEYLGAG---AERVLFDV-----------HPLN------EADT
MAV_1077|M.avium_104 DAEKLARWAELGVT---EVLFGLPD-----------KSED--------EV
** : . .
Mb3106c|M.bovis_AF2122/97 LRYLDKLQAELARLA-----------------------------
Rv3079c|M.tuberculosis_H37Rv LRYLDKLQAELARLA-----------------------------
MMAR_3559|M.marinum_M LRLLDEYAALVAQYHR----------------------------
MUL_2792|M.ulcerans_Agy99 PSFLREPLARVSCASRRKIAPRFAAQCDFASARGGRLRVGSRPR
MSMEG_1996|M.smegmatis_MC2_155 LRMLDDYATAVEKYRG----------------------------
Mflv_1017|M.gilvum_PYR-GCK LRLLDDYAEQVARYIG----------------------------
Mvan_5819|M.vanbaalenii_PYR-1 LRLLDGYAEQVARY------------------------------
TH_2750|M.thermoresistible__bu LRMLDQFAERVAGYRG----------------------------
MAB_4908c|M.abscessus_ATCC_199 LRALDRLTEVVARLT-----------------------------
MAV_1077|M.avium_104 AAYVERLAGKLAALV-----------------------------
: :