For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VISMSVVDGIRSLLDTTLDRALDRTIVGGYTRVGYQLRHSGWSGDPSPAALRGRTAMVTGANRGLGKAIA AGLAELGATVVLTVRDRDSGQRARDEIAAASGADVRVEVCDVSSLSVVRASATDLCTRVSRLDVLVHNAG LLPAARAETDEGHEITLATHVLGPILLTSLLVPMLAESDDPRVILMSSGGMYTQPLPIEDPEYRAGGYRG AVAYARTKRMQVAFTPLLARRWSGERIRVYSMHPGWADTPGVATSLPGFRAVIGPLLRTAEQGADTAVWL AATRPAPLSGTFWHDRRTRPEHYLPWTRYSDDDRQRLWRYCADAIGID
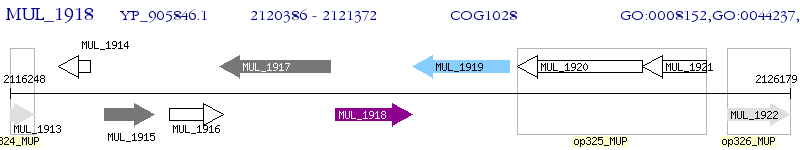
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1918 | - | - | 100% (328) | dehydrogenase/reductase |
| M. ulcerans Agy99 | MUL_1391 | - | 1e-27 | 32.01% (303) | short chain dehydrogenase |
| M. ulcerans Agy99 | MUL_1288 | - | 3e-21 | 32.51% (203) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0069 | - | 8e-31 | 35.54% (287) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0143 | - | 3e-26 | 33.81% (281) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv0068 | - | 6e-31 | 35.89% (287) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00315 | - | 1e-25 | 32.99% (291) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0646c | - | 7e-31 | 34.66% (277) | short chain dehydrogenase |
| M. marinum M | MMAR_1680 | - | 0.0 | 97.54% (325) | dehydrogenase/reductase |
| M. avium 104 | MAV_4710 | - | 2e-30 | 33.44% (308) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0876 | - | 1e-31 | 35.56% (284) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_4179 | - | 1e-30 | 35.05% (291) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_0753 | - | 4e-28 | 33.33% (291) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0069|M.bovis_AF2122/97 --------------------------------------MTKWTAADIPDQ
Rv0068|M.tuberculosis_H37Rv --------------------------------------MTKWTAADIPDQ
MLBr_00315|M.leprae_Br4923 --------------------------------------MAKWTTADIPDQ
MSMEG_0876|M.smegmatis_MC2_155 --------------------------------------MTKWTTARIPDQ
MAB_0646c|M.abscessus_ATCC_199 ------------------------------MSR----STTKWSTTDIPDQ
Mflv_0143|M.gilvum_PYR-GCK -------------------------------MT----DWTNWNVADIPDQ
TH_4179|M.thermoresistible__bu -------------------------------MT------GKWTARDVPDQ
Mvan_0753|M.vanbaalenii_PYR-1 -------------------------------MS------SKWTAADVPDQ
MAV_4710|M.avium_104 -------------------------------MAPHDKRHRDWSEADVGDQ
MUL_1918|M.ulcerans_Agy99 MISMSVVDGIRSLLDTTLDRALDRTIVGGYTRVGYQLRHSGWSGDPSPAA
MMAR_1680|M.marinum_M ---MSVVDGIRSLLDTTLDRALDRTIVGGYTRVGYQLRHSGWSDDPSPAA
*.
Mb0069|M.bovis_AF2122/97 T-GRTAVITGANTGLGFETAAALAAHGAHVVLAVRNLDKGKQAAARITEA
Rv0068|M.tuberculosis_H37Rv T-GRTAVITGANTGLGFETAAALAAHGAHVVLAVRNLDKGKQAAARITEA
MLBr_00315|M.leprae_Br4923 T-GRVAVITGANTGLGYQTALALAEHGAHVVLAVRNLDKGKDAAARITAT
MSMEG_0876|M.smegmatis_MC2_155 T-GRVAIVTGANTGLGLETAKALAAKGAHVVLAVRNLDKGKAAVDWIARS
MAB_0646c|M.abscessus_ATCC_199 T-GRIAIVTGANTGLGLETAKALAAHGAHVVLAVRNAEKGKAAAEAITAA
Mflv_0143|M.gilvum_PYR-GCK T-GRTAVITGANTGLGFETAKALAEKGARVVIAVRNTDKGAQAAARIRG-
TH_4179|M.thermoresistible__bu T-GRTAVITGANTGLGFETAKVLAEKGAHVVLAVRDPDKGRRAADRITAA
Mvan_0753|M.vanbaalenii_PYR-1 S-GRVAVVTGANSGIGYEAAAVLAGRGARVVVAVRNLDKGRQAVSRIRQL
MAV_4710|M.avium_104 S-GRVVVITGANTGIGYETAAVLAHRGAHVVLAVRDLEKGNAALSRIVAA
MUL_1918|M.ulcerans_Agy99 LRGRTAMVTGANRGLGKAIAAGLAELGATVVLTVRDRDSGQRARDEIAAA
MMAR_1680|M.marinum_M LRGRTAMVTGANRGLGKAIAAGLAELGATVVLTVRDRDSGQRARDEIAAA
** .::**** *:* * ** ** **::**: :.* * *
Mb0069|M.bovis_AF2122/97 TPGAEVELQELDLTSLASVRAAAAQLKSDHQRIDLLINNAGVMYTPRQTT
Rv0068|M.tuberculosis_H37Rv TPGAEVELQELDLTSLASVRAAAAQLKSDHQRIDLLINNAGVMYTPRQTT
MLBr_00315|M.leprae_Br4923 SAQNNVALQELDLASLESVRAAAKQLRSDYDHIDLLINNAGVMWTPKSTT
MSMEG_0876|M.smegmatis_MC2_155 APTADLELQQLDLGSLASVRAAADDLKGKFDRIDLLVNNAGVMWPPRQTT
MAB_0646c|M.abscessus_ATCC_199 HSNADVTLQSLDLSSLESVRRASDELKGRYDKIDLLINNAGVMWTEKSST
Mflv_0143|M.gilvum_PYR-GCK ----DVDVQELDLTSLSSIRTAAEALKARFDKIDLLINNAGVMTTPKGTT
TH_4179|M.thermoresistible__bu APHADVTVRQLDLTSLDNIRRAADDLRAGYPRIDLLINNAGVMYPPRQTT
Mvan_0753|M.vanbaalenii_PYR-1 HPGADVMLQELDLSSLASVRAAADDLRAAHPRIDLLINNAGVMYPPKQTT
MAV_4710|M.avium_104 SPNADVTLQQLDLASLASVRSAAEALRAAYPRIDLLINNAGVMWTPKQVT
MUL_1918|M.ulcerans_Agy99 SG-ADVRVEVCDVSSLSVVRASATDLCTRVSRLDVLVHNAGLLPAARAET
MMAR_1680|M.marinum_M SG-ADVRVEVCDVSSLSAVRAFATDLRTRVSRLDVLVHNAGLLPAARAET
:: :. *: ** :* : * ::*:*::***:: . : *
Mb0069|M.bovis_AF2122/97 ADGFEMQFGTNHLGHFALTGLLIDRLLPVAGSRVVTISSVGHRIRAAIHF
Rv0068|M.tuberculosis_H37Rv ADGFEMQFGTNHLGHFALTGLLIDRLLPVAGSRVVTISSVGHRIRAAIHF
MLBr_00315|M.leprae_Br4923 KDGFELQFGTNHLGHFAFTGLLLDRLLPIVGSRVITVSSLSHRLFADIHF
MSMEG_0876|M.smegmatis_MC2_155 ADGFELQFGTNHLGHFALTGLLLDRMLTVPGSRVVTVSSQGHRILAAIHF
MAB_0646c|M.abscessus_ATCC_199 ADGFELQFGTNHLGHYALTGLLLERLLPVEGSRVVTVSSIGHRIRADIHF
Mflv_0143|M.gilvum_PYR-GCK ADGFELQFGTNHLGHFALTGLLFDNILDIPGSRIVTVSSNGHKMGGAIHW
TH_4179|M.thermoresistible__bu RDGFELQFGTNHLGHFALTGQLLDNILPVDGSRVVTVASIAHRNMADIHF
Mvan_0753|M.vanbaalenii_PYR-1 SDGFELQFGTNHLGHFALTGLLLDRLLPVEGSRVVSVASIAHNIQADIHF
MAV_4710|M.avium_104 EDGFELQFGTNHLGHFALTGLLLDHLLGVRDSRVVTVSSLGHRLRAAIHF
MUL_1918|M.ulcerans_Agy99 DEGHEITLATHVLGPILLTSLLVPMLAESDDPRVILMSSGGMYTQPLPIE
MMAR_1680|M.marinum_M VEGHEITLATHVLGPILLTELLVPMLAESDDPRVILMSSGGMYTQPLPIE
:*.*: :.*: ** :* *. : ..*:: ::* .
Mb0069|M.bovis_AF2122/97 DDLQWERRYRRVAAYGQAKLANLLFTYELQRRLAPG--GTTIAVASHPGV
Rv0068|M.tuberculosis_H37Rv DDLQWERRYRRVAAYGQAKLANLLFTYELQRRLAPG--GTTIAVASHPGV
MLBr_00315|M.leprae_Br4923 NDLQWECNYNRVAAYGQSKLANLLFTYELQRRLATR--QTTIAVAAHPGG
MSMEG_0876|M.smegmatis_MC2_155 DDLQWERRYNRVAAYGQSKLANLLFTYELQRRLT-G--HQTTALAAHPGA
MAB_0646c|M.abscessus_ATCC_199 DDLQWERDYDRVAAYGQSKLANLLFTYELQRRLA-G--TNTVALAAHPGG
Mflv_0143|M.gilvum_PYR-GCK DDLQWERSYNRMGAYTQSKLANLLFTYELQRRLAPR--GKTIAVAAHPGT
TH_4179|M.thermoresistible__bu DDLQWERGYHRVAAYGQSKLANLMFAYELQRRLSAK-NAPTISVAAHPGV
Mvan_0753|M.vanbaalenii_PYR-1 DDLQWERSYNRVAAYGQSKLANLMFTYTLARRLAAK-GAPTIAVAAHPGI
MAV_4710|M.avium_104 DDLHWERRYDRVAAYGQSKLANLLFTYELQRRLAAAPDAKTIAVAAHPGG
MUL_1918|M.ulcerans_Agy99 DPEYRAGGYRGAVAYARTKRMQVAFTPLLARRWSGE---RIRVYSMHPGW
MMAR_1680|M.marinum_M DPEYRAGGYRGAVAYARTKRMQVAFTPLLARRWSGE---RIRVYSMHPGW
: * ** ::* :: *: * ** : : ***
Mb0069|M.bovis_AF2122/97 SNT-ELVRNMPRPLVAVAAILAP--LMQDAELGALPTLRAATDPAVRGGQ
Rv0068|M.tuberculosis_H37Rv SNT-EVVRNMPRPLVAVAAILAP--LMQDAELGALPTLRAATDPAVRGGQ
MLBr_00315|M.leprae_Br4923 SRT-ELTRTLPALIAPIFSVAELF-LTQDAATGALPTLRAATDAAVLGGQ
MSMEG_0876|M.smegmatis_MC2_155 SNT-ELARHLPG---ALERLVTP--LAQDAALGALPTLRAATDPGALGGQ
MAB_0646c|M.abscessus_ATCC_199 SNT-ELARNSPLWVRAVFDVVAPV-LVQGADMGALPTLRAATDPAALGGQ
Mflv_0143|M.gilvum_PYR-GCK STT-ELARNLPRPVERAFLAAAPVLFAQTADRGALPTLRAAADPGVLGGQ
TH_4179|M.thermoresistible__bu SNT-ELTRYIPGARLPGVSLLAG-LLTNSPAVGALATLRAATDPEVKGGQ
Mvan_0753|M.vanbaalenii_PYR-1 SNT-ELMRHIPGSQLPGFAWLAG-LVTNSPAVGSLATLRAATDPGVRGGQ
MAV_4710|M.avium_104 SNT-ELARHLPGIFRPVQAVLGP-VLFQSPAMGALPTLRAATDPAVQGAQ
MUL_1918|M.ulcerans_Agy99 ADTPGVATSLPG-----FRAVIGPLLRTAEQGADTAVWLAATRPAPLSGT
MMAR_1680|M.marinum_M ADTPGVATSLPG-----FRAVTGPLLRTAEQGADTAVWLAATRPAPLSGT
: * : * . . .. **: . ..
Mb0069|M.bovis_AF2122/97 YFGPDGFGEIRGYPKVVASSAQSHDEQLQRRLWAVSEELTGVVYPVG---
Rv0068|M.tuberculosis_H37Rv YFGPDGFGEIRGYPKVVASSAQSHDEQLQRRLWAVSEELTGVVYPVG---
MLBr_00315|M.leprae_Br4923 YFGPDGFAEIRGHPKVVASNGKSHDVDRQLRLWAVSEELTGVVYPVG---
MSMEG_0876|M.smegmatis_MC2_155 YFGPDGIGETRGYPKVVASSAQSHDADLQRRLWAVSEELTGVTFPVGEAL
MAB_0646c|M.abscessus_ATCC_199 YYGPDGFMEQRGNPKVVASSEQSYNLDLQRRLWSVSEELTDVVFPVK---
Mflv_0143|M.gilvum_PYR-GCK YYGPDGLGQQRGAPVVVASSAQSYDVDQQRRLWEISEELTKVTFPDL---
TH_4179|M.thermoresistible__bu YYGPDGFQEIRGHPVLVGSSAKSRDEDIQRRLWTVSEELTGVTFPV----
Mvan_0753|M.vanbaalenii_PYR-1 YYGPSGVRELVGHPVLVQSNRKSHDVDVQERLWTVSEELTGVSYDL----
MAV_4710|M.avium_104 YYGPDGFLEQRGRPKLVESSAQSHDEQLQRRLWAVSEELTGVHFPV----
MUL_1918|M.ulcerans_Agy99 FWH-----DRRTRPEHYLPWTRYSDDDRQ-RLWRYCADAIGID-------
MMAR_1680|M.marinum_M FWH-----DRRTRPEHYLPWTRFSDDDRQ-RLWRYCADAIGID-------
:: : * . : : : * *** . : :
Mb0069|M.bovis_AF2122/97 ---
Rv0068|M.tuberculosis_H37Rv ---
MLBr_00315|M.leprae_Br4923 ---
MSMEG_0876|M.smegmatis_MC2_155 QQA
MAB_0646c|M.abscessus_ATCC_199 ---
Mflv_0143|M.gilvum_PYR-GCK ---
TH_4179|M.thermoresistible__bu ---
Mvan_0753|M.vanbaalenii_PYR-1 ---
MAV_4710|M.avium_104 ---
MUL_1918|M.ulcerans_Agy99 ---
MMAR_1680|M.marinum_M ---