For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTLTAFEVSAAEASSDRSDRIPSTSVRPLTWRAALWSVMSVRWAGIALALFLTGLVAQLNGAAEPVWWTL YLACYLAGGWGSAWAGAQALRNRALDVDLLMIVAAIGAVAIGQIFDGALLIVIFATSGALDDVATKHTAD SVKGLLDLAPEQATVFDVDGNEQVVAASELVVGDRVVVRPGERIPADGAVLSGCSEVDQCSIAGESMPAA KGRGDEVFAGTVNGSGVLQLLVTRDPSQTVVARIVERVSEASATKAKTQLFIEKIEQRYSIGVVIATLAL IAIPLMLGTPVQVVLLRAMTFMIVASPCAVVLATMPPLLSAIADAGRHGVLVKSAIVVEHLADTGVVALD KTGTLTFGLPQLAIVEPLSADVGIKKLLQLAAAAEQSSEHPLGRAIVEEVRRRGITIPGAEDFRALPGRG VRASVGREFVEVCSPHSYRGAPLPELAPILEAGATAAIVLRNGVAIGVLGLTDQVRADAAPSVAALTALT SAPPVLLTGDNPCAAQRVAQHAGITDVRAALLPEQKVEAVQALQASGHRVLVVGDGVNDAPAMAAAHTSV AMGAGADPTLQTADGVTVRDELHTIPTLIGLARQARRVVTANLFIAGTFISVLVLWDLFGQLPLPLGVAG HEGSTVLVALNGMRLLTNRSWRAAASPVR
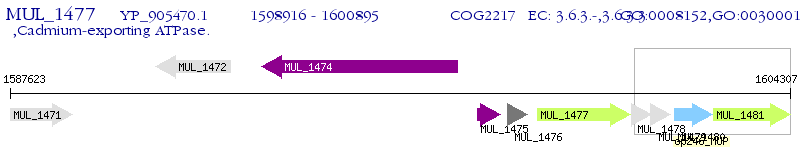
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1477 | ctpD | - | 100% (659) | cation transporter p-type ATPase D CtpD |
| M. ulcerans Agy99 | MUL_0423 | ctpV | 5e-73 | 33.44% (628) | metal cation transporter p-type ATPase, CtpV |
| M. ulcerans Agy99 | MUL_2616 | ctpC | 2e-66 | 30.84% (574) | metal cation-transporting p-type ATPase C CtpC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1504 | ctpD | 0.0 | 80.91% (660) | cation transporter P-type ATPase D |
| M. gilvum PYR-GCK | Mflv_2822 | - | 1e-102 | 37.22% (634) | heavy metal translocating P-type ATPase |
| M. tuberculosis H37Rv | Rv1469 | ctpD | 0.0 | 80.91% (660) | cation transporter P-type ATPase D |
| M. leprae Br4923 | MLBr_00747 | ctpC | 6e-65 | 30.72% (573) | putative cation transport ATPase |
| M. abscessus ATCC 19977 | MAB_4853c | - | 1e-95 | 35.46% (612) | cation-transporting ATPase G |
| M. marinum M | MMAR_2275 | ctpD | 0.0 | 99.09% (659) | cation transporter p-type ATPase D CtpD |
| M. avium 104 | MAV_5246 | - | 2e-71 | 35.93% (540) | copper-translocating P-type ATPase |
| M. smegmatis MC2 155 | MSMEG_5403 | cadA | 0.0 | 67.24% (638) | cadmium-translocating P-type ATPase |
| M. thermoresistible (build 8) | TH_2531 | - | 3e-72 | 33.79% (651) | PUTATIVE copper-translocating P-type ATPase |
| M. vanbaalenii PYR-1 | Mvan_3729 | - | 0.0 | 60.84% (641) | heavy metal translocating P-type ATPase |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_1477|M.ulcerans_Agy99 --------------------------------------------------
MMAR_2275|M.marinum_M --------------------------------------------------
Mb1504|M.bovis_AF2122/97 --------------------------------------------------
Rv1469|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_5403|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3729|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2822|M.gilvum_PYR-GCK MPPTRGFAQIGVDESYAHMRIIACMADRNGEKVGRLGADGAPTHRTLALD
MAB_4853c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_5246|M.avium_104 --------------------------------------------------
TH_2531|M.thermoresistible__bu --------------------------------------------------
MLBr_00747|M.leprae_Br4923 --------------------------------------------------
MUL_1477|M.ulcerans_Agy99 --------------------------------------------------
MMAR_2275|M.marinum_M --------------------------------------------------
Mb1504|M.bovis_AF2122/97 --------------------------------------------------
Rv1469|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_5403|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3729|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2822|M.gilvum_PYR-GCK LTDLLPDSDERCADRLTRGLVDDGIIEQAHVIDGDTPKPRLCVHYDAEAA
MAB_4853c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_5246|M.avium_104 --------------------------------------------MSTPPR
TH_2531|M.thermoresistible__bu --------------------------------------------MVGPAH
MLBr_00747|M.leprae_Br4923 ----------------------------------------MTLAMAEQIA
MUL_1477|M.ulcerans_Agy99 --------------------------------------------------
MMAR_2275|M.marinum_M --------------------------------------------------
Mb1504|M.bovis_AF2122/97 --------------------------------------------------
Rv1469|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_5403|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3729|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2822|M.gilvum_PYR-GCK SATEVQRRALEAADTITSMYGHLRWLTTGTHDDMAMRTALVDTLSTIPGV
MAB_4853c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_5246|M.avium_104 HIDEGTFPDHTASTARIELEITGMTCASCAARIEKKLNKLDGVTATVNYA
TH_2531|M.thermoresistible__bu HG--------TVAADGRAQHLTGR---------DHHRHGHG---------
MLBr_00747|M.leprae_Br4923 TADNPAFVVVSDAAGRMRIQIEWVRSNPRRAVTVEEAIAKCN--------
MUL_1477|M.ulcerans_Agy99 -------------------------------MTLTAFEVSAAEASSDRSD
MMAR_2275|M.marinum_M -------------------------------MTLTAFEVSAAEASSDRPD
Mb1504|M.bovis_AF2122/97 -------------------------------MTLTACEVTAAEAPFDR--
Rv1469|M.tuberculosis_H37Rv -------------------------------MTLTACEVTAAEAPFDR--
MSMEG_5403|M.smegmatis_MC2_155 ---------------------------------MTALYPAVEPAPAAR--
Mvan_3729|M.vanbaalenii_PYR-1 --------------------------MRVCAGIVVRVSADLQAADWERTT
Mflv_2822|M.gilvum_PYR-GCK VAAAATPDSVELEFERTTVTAQELVDVIAAQMAPSGSAADAEAAVAHD--
MAB_4853c|M.abscessus_ATCC_199 -------------------------------MSDACGCGHDEPATGDE--
MAV_5246|M.avium_104 TEKAAVSAPASYDPQTLITEIENAGYAAAVATPSPPRDDPELASLRRRLV
TH_2531|M.thermoresistible__bu ---------------------DHAGHVAEFRR--------LFWVMLLLAV
MLBr_00747|M.leprae_Br4923 -------------GVRVVHAYPRTGSVVVWYSPRCCDRQSILAAISGAAH
MUL_1477|M.ulcerans_Agy99 RIPSTSVRPLTWRAALWSVMSVRWAGIALALFLTG-LVAQLNGAAEPVWW
MMAR_2275|M.marinum_M RIPSTSVRPLTWRAALWSVMSVRWAGIALALFLTG-LVAQLNGAAEPVWW
Mb1504|M.bovis_AF2122/97 -VSKTIPHPLSWGAALWSVVSVRWATVALLLFLAG-LVAQLNGAPEAMWW
Rv1469|M.tuberculosis_H37Rv -VSKTIPHPLSWGAALWSVVSVRWATVALLLFLAG-LVAQLNGAPEAMWW
MSMEG_5403|M.smegmatis_MC2_155 -----PARPRS-GGWLWTVPSVRWAAAALALFLTG-LAAQLLGAPQAVVW
Mvan_3729|M.vanbaalenii_PYR-1 ATGEEVRGWRAPRSSAWSLPEVRWALIATVLFVVG-GLAQLVGAPPWLYW
Mflv_2822|M.gilvum_PYR-GCK --GDGADHQHGHGGISGERTELVFAGLAGALLLTGWLLAAFADTPRSAEV
MAB_4853c|M.abscessus_ATCC_199 --VDGEREPERLWEVS----ELRFAAVAGVVLLAGYVVG-WTGGSHPVEI
MAV_5246|M.avium_104 TAIALAG----PVIAVAMIPALQFQHWHWAALALTAPVVGWCGRPFHAAA
TH_2531|M.thermoresistible__bu PTVAFSG----MFAMILGYQVPRVPGAQWVSPVLGTVMYVWGGRPFLTGA
MLBr_00747|M.leprae_Br4923 VAAELIPTRAPHSSDIRNIEVLRMAIGAAALTLLGVRRYVFARPLLLPTT
MUL_1477|M.ulcerans_Agy99 TLYLACYLAGGWGSAWAGAQALRNRALD-------------------VDL
MMAR_2275|M.marinum_M TLYLACYLAGGWGSAWAGAQALRNRALD-------------------VDL
Mb1504|M.bovis_AF2122/97 TLYLACYLAGGWGSAWAGAQALRNKALD-------------------VDL
Rv1469|M.tuberculosis_H37Rv TLYLACYLAGGWGSAWAGAQALRNKALD-------------------VDL
MSMEG_5403|M.smegmatis_MC2_155 TLYLACYVVGGWEPAWVGVRALRNRTLD-------------------VDL
Mvan_3729|M.vanbaalenii_PYR-1 TLYLACYVTGGWEPGWAGLQALRDKTLD-------------------VDL
Mflv_2822|M.gilvum_PYR-GCK VVYGLAFFFGAFFTVQEAFASVRQGRFE-------------------IDF
MAB_4853c|M.abscessus_ATCC_199 GLYAAALAIGAYTFVPSTLRRLARGKIG-------------------VGT
MAV_5246|M.avium_104 WANLKHGVTTMDTLISIGTLAAFLWSLYALVLGAADRPGMRHDFELTVGH
TH_2531|M.thermoresistible__bu ISEIRSRTPGMMLLIGLALTVAFLSS-----------------------W
MLBr_00747|M.leprae_Br4923 SRLVASGVTIFTGYPFLRGALRFGKTG--------------------TDA
MUL_1477|M.ulcerans_Agy99 LMIVAAIGAVAIGQIFDGALLIVIFATSGALDDVATKHTADSVKGLLDLA
MMAR_2275|M.marinum_M LMIVAAIGAVAIGQIFDGALLIVIFATSGALDDVATKHTADSVKGLLDLA
Mb1504|M.bovis_AF2122/97 LMIAAAVGAVAIGQIFDGALLIVIFATSGALDDIATRHTAESVKGLLDLA
Rv1469|M.tuberculosis_H37Rv LMIAAAVGAVAIGQIFDGALLIVIFATSGALDDIATRHTAESVKGLLDLA
MSMEG_5403|M.smegmatis_MC2_155 LMIVAAIGAATIGQVFDGALLIVIFATSGALEDVATTRTERSVRGLLDLA
Mvan_3729|M.vanbaalenii_PYR-1 LMVAAAIGAAAIGQVFDGALLIVIFATSGALEAVATKRTEDSVRGLLDLA
Mflv_2822|M.gilvum_PYR-GCK LMLVSAAGAAALGEVAEGALLLFLFSVGHALEGYAMGRARRAIEALAELA
MAB_4853c|M.abscessus_ATCC_199 LMTIAAIGAVILGEVGEAAMLAFLFSISEGLEEYALARTRRGLRALLSLA
MAV_5246|M.avium_104 GAHVSHVAAPCHVYFEVAAGVTLFVLAGRYFERRSKRTAGAALRALLALG
TH_2531|M.thermoresistible__bu GASLGVLDHRLEFWWELALLIVIMLLG-HWIELRSLARTTSALDSLAALL
MLBr_00747|M.leprae_Br4923 LVSAATIASLILRENVVALAVLWLLNIGEYLQDLTLRRTRRAISALLSGT
. . : :. :: : : .: .*
MUL_1477|M.ulcerans_Agy99 PEQA-TVFDVDG---NEQVVAASELVVGDRVVVRPGERIPADGAVLSGCS
MMAR_2275|M.marinum_M PEQA-TVFDVDG---NEQVVAASELVVGDRVVVRPGERIPADGAVLSGCS
Mb1504|M.bovis_AF2122/97 PDQA-VVVQGDG---SERVVAASELVVGDRVVVRPGDRIPADGAVLSGAS
Rv1469|M.tuberculosis_H37Rv PDQA-VVVQGDG---SERVVAASELVVGDRVVVRPGDRIPADGAVLSGAS
MSMEG_5403|M.smegmatis_MC2_155 PEHA-TLLG-DG---SQRVVAAADLRPGDVIVVRPGERISADGTVIGGAS
Mvan_3729|M.vanbaalenii_PYR-1 PDTA-TRLTDSG---AEEVVDTAVLDVGDVLMIRPGERIGGDGTVVDGAS
Mflv_2822|M.gilvum_PYR-GCK PKTALVRRGGTG---DTVEVSVADLRIGDIVVVRPNMRLAADGFVVAGSS
MAB_4853c|M.abscessus_ATCC_199 PEEATVLRDGT-----ETTITATELQVGDTMLVRPGERVATDGTITAGRT
MAV_5246|M.avium_104 AKDVAV-LRAG----AETRIPIERLAVGDEFVVRPGERIATDGIVVAGSS
TH_2531|M.thermoresistible__bu PDTAER-VEQGPDGETVTVVAPADLRVGDTVIVRPGAGIPADGRIIDGVA
MLBr_00747|M.leprae_Br4923 QDTAWIRLTDGPQAGTEIQVPIGTVQIGDEVVVHEHVAIPVDGEVIDGEA
. . : : ** .::: : ** : * :
MUL_1477|M.ulcerans_Agy99 EVDQCSIAGESMPAAKGRG----------------DEVFAGTVNGSGVLQ
MMAR_2275|M.marinum_M EVDQCSITGESMPAAKGRG----------------DEVFAGTVNGSGVLQ
Mb1504|M.bovis_AF2122/97 DVDQRSITGESMPVAKARG----------------DEVFAGTVNGSGVLH
Rv1469|M.tuberculosis_H37Rv DVDQRSITGESMPVAKARG----------------DEVFAGTVNGSGVLH
MSMEG_5403|M.smegmatis_MC2_155 EVDQSSITGEPLPAAKDVG----------------DDVFAGTVNGSGALR
Mvan_3729|M.vanbaalenii_PYR-1 EVDQATITGEPLPVDKATG----------------DEVFAGTVNGTGTLR
Mflv_2822|M.gilvum_PYR-GCK SIDQAPVTGESVPVDKIPVPDVAAAAASPELIDAASRVFAGTINGAGAIE
MAB_4853c|M.abscessus_ATCC_199 ALNTSAITGESVPVEAGPG----------------DEVFAGSINGNGVLQ
MAV_5246|M.avium_104 AVDAAMLTGESVPVEVGVG----------------DGVTGGTVNAGGRLV
TH_2531|M.thermoresistible__bu DVDESMVTGESVPVRRGVG----------------DRVVAATVATDSGLR
MLBr_00747|M.leprae_Br4923 VVNQSAITGENLPVSVMAG----------------SHVHAGSVVVRGRLM
:: ::** :*. . * ..:: . :
MUL_1477|M.ulcerans_Agy99 LLVTRDPSQTVVARIVERVSEASATKAKTQLFIEKIEQRYSIGVVIATLA
MMAR_2275|M.marinum_M LLVTRDPSQTVVARIVELVSEASATKAKTQLFIEKIEQRYSIGVVIATLA
Mb1504|M.bovis_AF2122/97 LVVTRDPSQTVVARIVELVADASATKAKTQLFIEKIEQRYSLGMVAATLA
Rv1469|M.tuberculosis_H37Rv LVVTRDPSQTVVARIVELVADASATKAKTQLFIEKIEQRYSLGMVAATLA
MSMEG_5403|M.smegmatis_MC2_155 VEVTREPSQTVVARIVAMVTEASATKATTQLFIEKIEQRYSAGVVVATLA
Mvan_3729|M.vanbaalenii_PYR-1 VRVTRPAADTVIARIIAMVSEASASKAKMQLFIEKIEQRYSIGMVLATIA
Mflv_2822|M.gilvum_PYR-GCK IQVTRLASDSTLARVVRLVAEAQTKTTSTQRFTDRFQRIFVPVVLVGVVL
MAB_4853c|M.abscessus_ATCC_199 VRVSATAADNSLARIVAIVEAEQSRKGASQRLADRIAKPLVPAIMITAAA
MAV_5246|M.avium_104 VRATGIGDDTQLARMAQLVERAQSGKADAQRLADRVSGVFVPVVLLLAVA
TH_2531|M.thermoresistible__bu ITVTAVGDDTALAGIRRMVAEAQSSSSRAQRIADTAAGWLFWFALGAALL
MLBr_00747|M.leprae_Br4923 VRASAVGKHTTIGRIVTRVEEAQHDRAPIQTVGENFSRCFVPTSFVVSAI
: .: .. :. : * . * . : .
MUL_1477|M.ulcerans_Agy99 LIAIPLMLGTPVQVVLLRAMTFMIVASPCAVVLATMPPLLSAIADAGRHG
MMAR_2275|M.marinum_M LIAIPLMLGTPVQVVLLRAMTFMIVASPCAVVLATMPPLLSAIANAGRHG
Mb1504|M.bovis_AF2122/97 LIVIPLMFGADLRPVLLRAMTFMIVASPCAVVLATMPPLLSAIANAGRHG
Rv1469|M.tuberculosis_H37Rv LIVIPLMFGADLRPVLLRAMTFMIVASPCAVVLATMPPLLSAIANAGRHG
MSMEG_5403|M.smegmatis_MC2_155 LLTVPLMFGADLRSTLLRAMTFMIVASPCAVVLATMPPLLSAIANASRHG
Mvan_3729|M.vanbaalenii_PYR-1 LFTIPLVLGADLQPTLLRAMTFMIVASPCALVLSTMPPLLSAIANAGRHG
Mflv_2822|M.gilvum_PYR-GCK LLFAGFVVDEPFTDTVYRALAVLVAASPCALAIATPSAVLSAVARAARAG
MAB_4853c|M.abscessus_ATCC_199 IAVLGSVFGDPLV-WIERALVVLVAASPCALAISVPVTVVAAIGAASKLG
MAV_5246|M.avium_104 TLAGWLAAGGTLATALTAAVAVLIIACPCALGLATPTALLVGTGRAAQLG
TH_2531|M.thermoresistible__bu TALVWAVLH-RPDFAVVRTITVLVIACPHALGLAIPLVVAIATERAARGG
MLBr_00747|M.leprae_Br4923 TLAITKDVR--------RTMTVLLIACPCAVGLATPTAISAAIGNGARRG
::..:: *.* *: :: : . ..: *
MUL_1477|M.ulcerans_Agy99 VLVKSAIVVEHLADTGVVALDKTGTLTFGLPQLAIVEPLSADVG-IKKLL
MMAR_2275|M.marinum_M VLVKSAIVVEHLADTGVVALDKTGTLTFGLPQLAIVEPLSVDVG-IKKLL
Mb1504|M.bovis_AF2122/97 VLVKSAVVVERLADTSIVALDKTGTLTRGIPRLASVAPLDPNVVDARRLL
Rv1469|M.tuberculosis_H37Rv VLVKSAVVVERLADTSIVALDKTGTLTRGIPRLASVAPLDPNVVDARRLL
MSMEG_5403|M.smegmatis_MC2_155 VLVKSAVAMERLADTDVVVLDKTGTLTAGEPVISRVTVLIDGAD----VL
Mvan_3729|M.vanbaalenii_PYR-1 VLVKSAVVLEKLSTVTHVAFDKTGTLTEGSPRLVHIRPVPDAAVDDAELL
Mflv_2822|M.gilvum_PYR-GCK ILMKGGAALEELGRVNVLAFDKTGTLTEGRPRIADVCATADSDD--AELL
MAB_4853c|M.abscessus_ATCC_199 VLVKGGAALEALGVVRTIALDKTGTLTANRPTVIEVAATAGATG--ERVL
MAV_5246|M.avium_104 VLIKGPEVLETTRAVDTVVLDKTGTVTTGAMTVLDVVAADGTDR--ATLL
TH_2531|M.thermoresistible__bu VLIKDRIALENMRTVDVVLFDKTGTLTKGQPAVTGITAAGAHSE--EDVL
MLBr_00747|M.leprae_Br4923 ILIKGGSHLEQAGRVDAILFDKTGTLTVGRPVVTNIVAMHKDWSP-EQVL
:*:*. :* . : :*****:* . : : :*
MUL_1477|M.ulcerans_Agy99 QLAAAAEQSSEHPLGRAIVEEV--RRRGITIPGAEDFRALPGRGVRASVG
MMAR_2275|M.marinum_M QLAAAAEQSSEHPLGRAIVEEV--RRRGITIPGAEDFRALPGRGVRASVG
Mb1504|M.bovis_AF2122/97 QLAAAAEQSSEHPLGRAIVAEA--RRRGIAIPPAKDFRAVPGCGVHALVG
Rv1469|M.tuberculosis_H37Rv QLAAAAEQSSEHPLGRAIVAEA--RRRGIAIPPAKDFRAVPGCGVHALVG
MSMEG_5403|M.smegmatis_MC2_155 GMAAAAEQFSEHPLGRAIVAAA--RGR--VVPEAGDFTALPGRGVRARVA
Mvan_3729|M.vanbaalenii_PYR-1 ALAAAAENPSEHPLARAIVAAA--RDAGLTLQPVEDFGSTPGHGVRARIG
Mflv_2822|M.gilvum_PYR-GCK RIAVAVEEQSDHPLARAIVRDGRERLAGAVTPRATDVRAVIGRGIVASVE
MAB_4853c|M.abscessus_ATCC_199 SAAAALESRSEHPLAAAILAA-----AGSVDP-ADDVEAVTSAGLTGRRG
MAV_5246|M.avium_104 RYAGALEAASEHPIAHAIARDA--KAELGPLPTPTGFRAVGGGGVHGRVD
TH_2531|M.thermoresistible__bu RLAAAAEAPSEHPLARALVAAA--QRHGTDLPPATDFASTPGLGVRARVA
MLBr_00747|M.leprae_Br4923 AYAASSEIHSRHPLAEAVIRST--EERHISIPPHEECEVLVGLGMRTWAD
* : * * **:. *: . *:
MUL_1477|M.ulcerans_Agy99 REFVEVCSPHSYRG-------APLPELAPILEAGATAAIVLRNGVAIGVL
MMAR_2275|M.marinum_M REFVEVCSPHSYRG-------APLPELAPILEAGATAAIVLRNGVAIGVL
Mb1504|M.bovis_AF2122/97 NDFVEIASPQSYRG-------APLAELAPLLSAGATAAIVLLDGVAIGVL
Rv1469|M.tuberculosis_H37Rv NDFVEIASPQSYRG-------APLAELAPLLSAGATAAIVLLDGVAIGVL
MSMEG_5403|M.smegmatis_MC2_155 GHVVEVVSPAAYAGE----NAAVREHCAAIENDGGTAVVVLEDGLPVGVI
Mvan_3729|M.vanbaalenii_PYR-1 EDLVEVGNPTLLPAV----REIAHEMVDDLEQTGHTAVIVRLNGTVVGVL
Mflv_2822|M.gilvum_PYR-GCK GVEVCIGKTELFTDAAQPPPAELAAEVNRLEQAGRTTMLVRAGDRWLGAI
MAB_4853c|M.abscessus_ATCC_199 GHTLRLGRPGWID------PGSLAPDVARMQQGGATAVLVEEDGQLLGAI
MAV_5246|M.avium_104 GHAVAIGRPSWLAERGLRPDAALAAAAARAEHDGKTVVAVGWDGRARGIL
TH_2531|M.thermoresistible__bu GRTVRVGGPRLLAETGQTAPAGSEGWRAR----GSTVLYVLVDDMIAGAV
MLBr_00747|M.leprae_Br4923 GRTLLLGSPSLLCAEKVKVSKTASEWVDKLRHQTETPLLFAVDGTLVGLI
: : . * . .. * :
MUL_1477|M.ulcerans_Agy99 GLTDQVRADAAPSVAALTALTSAPPVLLTGDNPCAAQRVAQHAGITDVRA
MMAR_2275|M.marinum_M GLTDQVRADAAPSVAALTALTSAPPVLLTGDNPCAAQRVAQHAGITDVRA
Mb1504|M.bovis_AF2122/97 GLTDQLRPDAVESVAAMAALTAAPPVLLTGDNGRAAWRVARNAGITDVRA
Rv1469|M.tuberculosis_H37Rv GLTDQLRPDAVESVAAMAALTAAPPVLLTGDNGRAAWRVARNAGITDVRA
MSMEG_5403|M.smegmatis_MC2_155 GLADRLRPDAPAAVMQLAQLTKHPPMLLTGDNRRAAGRLAEEAGIADVHA
Mvan_3729|M.vanbaalenii_PYR-1 GLTDRVRDAAADMVAALTELTGAQPILLTGDNPRAAAALAAQVGITDVRA
Mflv_2822|M.gilvum_PYR-GCK GLMDLPRPEASAVIARLAALGVKNTVMLSGDNQRVADAVAAEVGVAHARG
MAB_4853c|M.abscessus_ATCC_199 AVRDELRPEAAHVIAALHRSGYR-VAMLTGDNAATATTLARQVGIDTVHA
MAV_5246|M.avium_104 ALADTVKPSSAAAVRQFTRLGLT-PILLTGDNHTVARRTAGELGIGEVIS
TH_2531|M.thermoresistible__bu ALADEIRPESRPTVDALHRQGID-VALITGDARSVAESVAAELGIDRVLA
MLBr_00747|M.leprae_Br4923 SLRDEVRPEAAEVLTKLRASGVRRIVMLTGDHPDIAKAVATELGIDEWRA
.: * : : : : :::** * * . *: .
MUL_1477|M.ulcerans_Agy99 ALLPEQKVEAVQALQASGHRVLVVGDGVNDAPAMAAAHTSVAMG-AGADP
MMAR_2275|M.marinum_M ALLPEQKVEAVQALQASGHRVLVVGDGVNDAPAMAAAHTSVAMG-AGADL
Mb1504|M.bovis_AF2122/97 ALLPEQKVEVVRNLQAGGHQVLLVGDGVNDAPAMAAARAAVAMG-AGADL
Rv1469|M.tuberculosis_H37Rv ALLPEQKVEVVRNLQAGGHQVLLVGDGVNDAPAMAAARAAVAMG-AGADL
MSMEG_5403|M.smegmatis_MC2_155 ELLPDGKAAAVQKLQRDNTHVLVVGDGVNDAPAMAAAHTSIAMGRAGADL
Mvan_3729|M.vanbaalenii_PYR-1 GLLPQDKVEAVRELEAAGAKVMLVGDGVNDAPAMALATVGVAKGRTGSDL
Mflv_2822|M.gilvum_PYR-GCK DLMPEDKVAQIAALRERHGRVGMVGDGVNDAPAMAGASVGIAMGAAGSDV
MAB_4853c|M.abscessus_ATCC_199 DLRPEDKARIITELRQQHP-TAMVGDGVNDAPALATADLGIAMGAMGTDV
MAV_5246|M.avium_104 GALPADKVEAVKRLQSAGRVVAMVGDGVNDAAALAAADLGLAMG-TGTDV
TH_2531|M.thermoresistible__bu DVPPEHKAAKVVELQREGRRVAMVGDGVNDAPALARADVGIAIG-AGTDV
MLBr_00747|M.leprae_Br4923 EVMPEDKLKVVRDLQNEGYVVGMVGDGVNDAPALAAADIGIAMGLAGTDV
* * : *. . :********.*:* * .:* * *:*
MUL_1477|M.ulcerans_Agy99 TLQTADGVTVRDELHTIPTLIGLARQARRVVTANLFIAGTFISVLVLWDL
MMAR_2275|M.marinum_M TLQTADGVTVRDELHTIPTLIGLARQARRVVTANLFIAGTFISVLVLWDL
Mb1504|M.bovis_AF2122/97 TLQTADGVTIRDELHTIPTIIGLARQARRVVTVNLAIAATFIAVLVLWDL
Rv1469|M.tuberculosis_H37Rv TLQTADGVTIRDELHTIPTIIGLARQARRVVTVNLAIAATFIAVLVLWDL
MSMEG_5403|M.smegmatis_MC2_155 TVQTADVVTIRDELATVPAVIALARRARRVVIANLVMAGAAITTLVLWDL
Mvan_3729|M.vanbaalenii_PYR-1 ALATADAVITRDDLSTIPAVLALARRARRLVIANLTIAATLITVLVAWDL
Mflv_2822|M.gilvum_PYR-GCK ALETADVALMADDLRALPFAISLSRRSSRVIKQNLWASLGIVAVLIPATI
MAB_4853c|M.abscessus_ATCC_199 AIETADVALMGEDLRHLTQALAHARRARTIMLQNVGLSLALIAVLIPLAL
MAV_5246|M.avium_104 AIEAADVTVVRGDLRAAVDAIRLSRRTLATIKTNLVWAFGYNLAAIPLAA
TH_2531|M.thermoresistible__bu AIASAGVVLAGSDPRSVLSVLQLSRAAYRKMKQNLWWAAGYNLIAVPLAA
MLBr_00747|M.leprae_Br4923 AVETADVALANDDLNRLLDVRDLGGRAVEVIRENYGMSIAVNAAGLFIGA
:: :*. . : . : : * : :
MUL_1477|M.ulcerans_Agy99 FGQLPLPLGVAGHEGSTVLVALNGMRL-----LTNRSWRAAASPVR----
MMAR_2275|M.marinum_M FGQLPLPLGVAGHEGSTVLVALNGMRL-----LTNRSWRAAASPVR----
Mb1504|M.bovis_AF2122/97 FGQLPLPLGVVGHEGSTVLVALNGMRL-----LTNRSWRAAASAAR----
Rv1469|M.tuberculosis_H37Rv FGQLPLPLGVVGHEGSTVLVALNGMRL-----LTNRSWRAAASAAR----
MSMEG_5403|M.smegmatis_MC2_155 FGQLPLPLGVAGHEGSTILVALNGLRL-----LSNRAWISPGATPT----
Mvan_3729|M.vanbaalenii_PYR-1 IGHLPLPLGVAGHEGSTIVVGLNGLRL-----LRDAAWPSTRGTKATAKT
Mflv_2822|M.gilvum_PYR-GCK FG-LGIGPAVLIHEGSTLIVVANALRL-----LGMPMQSVTPAAPRVERA
MAB_4853c|M.abscessus_ATCC_199 LGILGLAAVVAVHELAEIVVIANGVRAGRTQPLTAPEHANTPAAQLAQQE
MAV_5246|M.avium_104 -----LGMLNPMLAGAA-MALSSVLVVGNS-----LRLRSFASIIPGA--
TH_2531|M.thermoresistible__bu GVLAPAGFVMPMSVGAILMSLSTVVVALNAQLLRRLDLRPEQSLAAARR-
MLBr_00747|M.leprae_Br4923 G-----GALSPVLAAVLHNASSVAVVANSS---RLIRYRLD---------
:
MUL_1477|M.ulcerans_Agy99 ----
MMAR_2275|M.marinum_M ----
Mb1504|M.bovis_AF2122/97 ----
Rv1469|M.tuberculosis_H37Rv ----
MSMEG_5403|M.smegmatis_MC2_155 ----
Mvan_3729|M.vanbaalenii_PYR-1 GK--
Mflv_2822|M.gilvum_PYR-GCK EETP
MAB_4853c|M.abscessus_ATCC_199 ----
MAV_5246|M.avium_104 ----
TH_2531|M.thermoresistible__bu ----
MLBr_00747|M.leprae_Br4923 ----