For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RNFQRRGAAIATFAFAFAAAPVIGLGAPAISRADDHNYMSGWTWDNVAQQCVISPVPATTAPAQNGPPPP GTPGGVYGPGGWGGPGGPGGPPASGEPYGPYGPQGGAPGTPGWPGGIAGPTAAGGHAGMGGIGGMDGMGG MGRH*
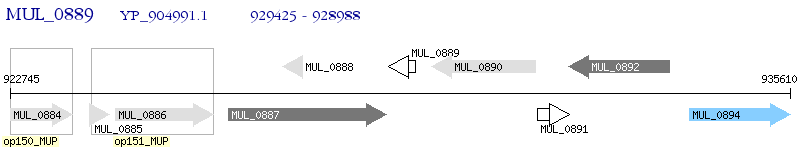
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0889 | - | - | 100% (145) | hypothetical protein MUL_0889 |
| M. ulcerans Agy99 | MUL_4367 | - | 2e-11 | 44.59% (74) | PE-PGRS family protein |
| M. ulcerans Agy99 | MUL_0391 | - | 2e-11 | 46.15% (78) | PE-PGRS family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1485c | PE_PGRS27 | 3e-14 | 47.56% (82) | PE-PGRS family protein |
| M. gilvum PYR-GCK | Mflv_5091 | - | 3e-12 | 46.84% (79) | triple helix repeat-containing collagen |
| M. tuberculosis H37Rv | Rv1452c | PE_PGRS28 | 1e-12 | 47.30% (74) | PE-PGRS family protein |
| M. leprae Br4923 | MLBr_01556 | infB | 1e-07 | 35.42% (96) | translation initiation factor IF-2 |
| M. abscessus ATCC 19977 | MAB_3220 | - | 6e-12 | 45.24% (84) | hypothetical protein MAB_3220 |
| M. marinum M | MMAR_1128 | - | 1e-86 | 97.24% (145) | hypothetical protein MMAR_1128 |
| M. avium 104 | MAV_2780 | - | 2e-11 | 44.44% (90) | hypothetical protein MAV_2780 |
| M. smegmatis MC2 155 | MSMEG_3545 | - | 2e-11 | 46.59% (88) | hypothetical protein MSMEG_3545 |
| M. thermoresistible (build 8) | TH_3810 | - | 7e-09 | 41.43% (70) | PUTATIVE - |
| M. vanbaalenii PYR-1 | Mvan_1264 | - | 3e-12 | 50.00% (72) | triple helix repeat-containing collagen |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_0889|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1128|M.marinum_M --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 ---------------------MSLVIVAPETVAAAALDVARIGSSIGVAN
Rv1452c|M.tuberculosis_H37Rv ---------------------MSLVIVTPETVAAAASDVARIGSSIGVAN
Mflv_5091|M.gilvum_PYR-GCK -------------------------------MVTFGGGATPTPTSLGAAA
Mvan_1264|M.vanbaalenii_PYR-1 MMFLAPPGGPGPISAAIQLTSTGSEATEPWYVVSFGGSGAPTPNNLAASL
TH_3810|M.thermoresistible__bu --------------------------------------------------
MSMEG_3545|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3220|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2780|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 ------------------------------------MAGKARVHELAKEL
MUL_0889|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1128|M.marinum_M --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 SAAAGSTTSVLAAGADEVSAAIATLFGSHAREYQAISTQVAAFHDRFAQT
Rv1452c|M.tuberculosis_H37Rv SAAAGSTTSVLAAGADEVSAAIATLFGSHAREYQAISTQVAAFHDRFAQT
Mflv_5091|M.gilvum_PYR-GCK VDLSGTSCGLICNGQDGTGQNGGILFGNGGSGWNSDVVGMDGGNGGNGGL
Mvan_1264|M.vanbaalenii_PYR-1 VDLSGTSCGLICNGQDGTGQNGGILFGNGGSGWNSEIAGVSGGNGGNAGL
TH_3810|M.thermoresistible__bu --------------------------------------------------
MSMEG_3545|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3220|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2780|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 GVTSKEVLARLNEQGEFVKSASSTVEAPVARRLRESFGGIKPAADKGAEQ
MUL_0889|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1128|M.marinum_M --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 LSAAVGSYVSAEATNAAPLATLEHNVLNALNAPTQALLGRPLIGDGAAGA
Rv1452c|M.tuberculosis_H37Rv LSAAVGSYVSAEATNAAPLATLEHNVLNALNAPTQALLGRPLIGDGAAGA
Mflv_5091|M.gilvum_PYR-GCK FGGNGGNGGNGLGGGNG---------GNGGNAGAFALFGN--GGNGGNGG
Mvan_1264|M.vanbaalenii_PYR-1 FGGNGGNGGNGLGSGDG---------GNGGDAGLFSLFGR--GGNGGNGG
TH_3810|M.thermoresistible__bu VDGDGGS---------------------GGRTGLFSKLSR--GGDGGDG-
MSMEG_3545|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3220|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2780|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 VATKAQAKRLG----------------ESLDQTLDRALDKAVAGNGATTA
MUL_0889|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1128|M.marinum_M --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 PGTGQAGGAGGILWGNGGAGGSGAPGQVGGAGG--AAGLFGTGGAGGAGG
Rv1452c|M.tuberculosis_H37Rv PGTGQAGGAGGILWGNGGAGGSGAPGQVGGAGG--AAGLFGTGGAGGAGG
Mflv_5091|M.gilvum_PYR-GCK VGVVGATGAVGAPGVAGATGATGVTSGVVGGNGEDAVGYGATGAAGAAGA
Mvan_1264|M.vanbaalenii_PYR-1 VGIAGATGAVGAPGVAGATGATGVTSNVTGGNGADAVGYGATGAAGAAGA
TH_3810|M.thermoresistible__bu -GLGGNGGHG---------GGTGIISLLS---------LAGNGGNGGNGY
MSMEG_3545|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3220|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2780|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 APVQVDHSAAVVPIVAGEGPSTAHREELAPPAGQPSEQPGVPLPGQQGTP
MUL_0889|M.ulcerans_Agy99 --------------------------MRNFQRRGAAIATFAFAFAAAPVI
MMAR_1128|M.marinum_M --------------------------MRNFQRRGAAIAAFAFAFAAAPVI
Mb1485c|M.bovis_AF2122/97 AGAAGGAGGSG-GWLLGNGGVGGAGGQSLLGGATGGAGGNAGLFGVGGTG
Rv1452c|M.tuberculosis_H37Rv AGAAGGAGGSG-GWLLGNGGVGGAGGQSLLGGATGGAGGNAGLFGVGGTG
Mflv_5091|M.gilvum_PYR-GCK AGAAGAAGAVGSAGAAGEAGETGTWGNAGLPGKAGDAGG-AGAVGAAGAA
Mvan_1264|M.vanbaalenii_PYR-1 AGAAGKAGAVGSAGTAGSAGAPGVWGNAGLPGAAGAAGG-AGAIGTAGVA
TH_3810|M.thermoresistible__bu R----------------------TTDNNALDGGHGGNGGNVGFFSLFNLG
MSMEG_3545|M.smegmatis_MC2_155 --------------------------MMIVKTTTTRFVAAALTVGALGFG
MAB_3220|M.abscessus_ATCC_1997 ---------MQDSRQRKKQDQRSPTHLTREHPMKTRTVKPAKVFAGCAIA
MAV_2780|M.avium_104 ------------------------------------------MFGGAAML
MLBr_01556|M.leprae_Br4923 AAPHPGHPGMPTGPHPGPAPKPGGRPPRVGNNPFSSAQSVARPIPRPPAP
.
MUL_0889|M.ulcerans_Agy99 G-------------------------------------------------
MMAR_1128|M.marinum_M G-------------------------------------------------
Mb1485c|M.bovis_AF2122/97 GPGGPGGPGGPGGPGGPGGVGGTGGAGGLGGTLYGAGGHGGAGGPGPIGG
Rv1452c|M.tuberculosis_H37Rv GPGGPG---------GPGGVGGTGGAGGLGGTLYGAGGHGGAGGPGPIGG
Mflv_5091|M.gilvum_PYR-GCK GTVGTAGATGG---TGTGANGGAGGVGGVGGTG-AAGAAGGAGGLGETGT
Mvan_1264|M.vanbaalenii_PYR-1 GAAGTAGATGG---TGVGGNGGAGQAGGIGGTG-AAGAAGQAGTAGETGV
TH_3810|M.thermoresistible__bu G----------------------------------AGGWGGSG-------
MSMEG_3545|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3220|M.abscessus_ATCC_1997 LG------------------------------------------------
MAV_2780|M.avium_104 V-------------------------------------------------
MLBr_01556|M.leprae_Br4923 RPSASP--------------------------------------------
MUL_0889|M.ulcerans_Agy99 ------------------------------------------LGAPAISR
MMAR_1128|M.marinum_M ------------------------------------------LGAPAISR
Mb1485c|M.bovis_AF2122/97 VGGHGGVGGAAGLLGVGGHGGAGGHGAAGIAGAAGKGGIFP-DGSNGGAG
Rv1452c|M.tuberculosis_H37Rv VGGHGGVGGAAGLLGVGGHGGAGGHGAEGVAGAAG-EDLSP-HGTSGGVG
Mflv_5091|M.gilvum_PYR-GCK TGGAGGVGGQGTAGGTGGVGGIGGAGSTGAVGGVGGKGIQGGQGLQGGKG
Mvan_1264|M.vanbaalenii_PYR-1 TGGVGGIGGQGTEGGTGGVGGVGGVGATGGAGGVGGKGIQGGQGLQGGQG
TH_3810|M.thermoresistible__bu VDGVDVVGGNP----DGGTPGAHGESHT--------------PGVYTDRQ
MSMEG_3545|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3220|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2780|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 ------------------------------------------SSMSPRPG
MUL_0889|M.ulcerans_Agy99 ADDHN-------------YMSGWTWDNVAQQCVISPVPATTAPAQNGPPP
MMAR_1128|M.marinum_M ADDHN-------------CMSGWTWDNVAQQCVISPVPATTAPAQNGPPP
Mb1485c|M.bovis_AF2122/97 GDAGDGGTGGRGGWLAGAGGAGGDGGIGGTGGAGGAGFSRALIVAGDNGG
Rv1452c|M.tuberculosis_H37Rv GDAGDGGTGGRGGWLAGAGGAGGAGGVGGTGGAGGAGFSRALIVAGDNGG
Mflv_5091|M.gilvum_PYR-GCK GDGGAGGKGGDADSMGSAGATGGKGGNGGNGGRGGLFGKGGNAGNGADGG
Mvan_1264|M.vanbaalenii_PYR-1 GDGGTGGTGGDATSMGEAGATGGKGGNGGNGGRGSLFGRGGNAGNGGNGG
TH_3810|M.thermoresistible__bu GEDGEDGTG---------TGIGGRGGNGGSSSGTHPMAEDGEHVKALDGR
MSMEG_3545|M.smegmatis_MC2_155 -----------------ATAFGTPVASAGPAAPVPLKPGDGGCFPFCDGG
MAB_3220|M.abscessus_ATCC_1997 ---------------AALAGCGTNNDGGPTSTSTTTTTTTTTTTTITPAP
MAV_2780|M.avium_104 ----------------LAVGCGGGHKGGPSTTSTTTPTTTPSTSVVPSTT
MLBr_01556|M.leprae_Br4923 GAVGGGGPRPPRTGVPRPGGGRPGAPVGGRSDAGGGNYRGGGVGALPGGG
MUL_0889|M.ulcerans_Agy99 PGTPGGVYGPGGWGGPGGPG---------GPPASGEPYGPYGPQG-----
MMAR_1128|M.marinum_M PGTPGGVYGPGGWGGPGGPG---------GPPAPGEPYGPYGPQV-----
Mb1485c|M.bovis_AF2122/97 DGGNGGMGGAGGAGGPGGAGGLISLLGGQGAGGDGGDGGAGGVGGDRGAG
Rv1452c|M.tuberculosis_H37Rv DGGNGGMGGAGGAGGPGGAGGLISLLGGQGAGGAGGTGGAGGVGGDRGAG
Mflv_5091|M.gilvum_PYR-GCK TGGAGGAGGTGGTGGAGGTGGTGGTGGIGGAGSTGGTGGTGGTGGTGGTG
Mvan_1264|M.vanbaalenii_PYR-1 TGGNGGNGGTGGTGGNGGTGGTGGTGGIGGAGSTGGTGGTGGTGGTGGTG
TH_3810|M.thermoresistible__bu PGLDGGNGSHGGNGGHGGRG------GFLGIGGSGGWGGHGGNGGD----
MSMEG_3545|M.smegmatis_MC2_155 PGKPGGPGGPG-HGGPGGPGGP----GHGGPGGPGGHEGPGGPGRHDG--
MAB_3220|M.abscessus_ATCC_1997 SGAPEGTPAPGPTGGGGPDGG--------SGAIPGGPTGGGGPEG-----
MAV_2780|M.avium_104 PGG-GGTVPGGPTGGGGPGGG--------GGSIPGGPTGGGGPGG-----
MLBr_01556|M.leprae_Br4923 SGGFRGRPGGGGHGGGGRPGQRGGAAGAFGRPGGAPRRGRKSKRQKRQEY
* * * ** * * . . * .
MUL_0889|M.ulcerans_Agy99 --------GAPGTPGWPGGIAGPTAAGGHAG-------------------
MMAR_1128|M.marinum_M --------GAPGTPGWPGGIAGPTAAGGHAG-------------------
Mb1485c|M.bovis_AF2122/97 ANGNQAVNAGAGGAGGHGGDPGAGGAGGTGGAGSTIGAHGAAGASPTSGG
Rv1452c|M.tuberculosis_H37Rv GPGNQAFNAGAGGAGGHGGDPGAGGAGGTGGAGSITGAQGAIGATPTSGG
Mflv_5091|M.gilvum_PYR-GCK GTGGTGGTGGTGGTGGKGGTGGTGGAGGTGGDGG-TGGTGGTGGTGGTGG
Mvan_1264|M.vanbaalenii_PYR-1 GTGGTGGTGGTGGTGGKGGTGGTGGDGGTGGTGG-TGGTGGTGGTGGTGG
TH_3810|M.thermoresistible__bu -------TDADGGNGGDGGNGGSAGRGGTG---------GLRGVFGINGG
MSMEG_3545|M.smegmatis_MC2_155 --------PGPGGPGGPGRHDGPGGPGGPGWHDP----------------
MAB_3220|M.abscessus_ATCC_1997 --------GSGSIPGGPTGGGGPEGGGGHIPGVG----------------
MAV_2780|M.avium_104 --------GGGSIPGGPTGGGGPGGGGGSIPGVG----------------
MLBr_01556|M.leprae_Br4923 DSMQAPVVGGVRLPHGNGETIRLARGASLSDFAEKIDANPAALVQALFNL
. ..
MUL_0889|M.ulcerans_Agy99 ----------------------MGGIGGMDGMGGMGRH------------
MMAR_1128|M.marinum_M ----------------------MGGIGGMDGMGGMGRH------------
Mb1485c|M.bovis_AF2122/97 NGGAGGNGAHFSSGGKAGGNGGAGGAGGLVGNGGAGGAGGNGAPGAPPSG
Rv1452c|M.tuberculosis_H37Rv NGGAGGNGANATTAGTNGANGGPGGHGGLVGNGGAGGNGANGAAGTNASD
Mflv_5091|M.gilvum_PYR-GCK TGGTGGAGDGTGGTGGTGGTGGQGGQGGQGGTGGTGGQGGLGGGGGTGGQ
Mvan_1264|M.vanbaalenii_PYR-1 TGGTGGAGDGLGGTGGTGGTGGFGGQGGQGGTGGTGGQGGLGGVGGTGGT
TH_3810|M.thermoresistible__bu NGRAG-----------------LGGRGXXXXGGGVGAAVR----------
MSMEG_3545|M.smegmatis_MC2_155 ------------------------GHGPDKGPWWAN--------------
MAB_3220|M.abscessus_ATCC_1997 -----------------------GGHGGPGGGGGCVDNVGCVSIP-----
MAV_2780|M.avium_104 -----------------------GGGGGPGGGGGCVGNV-CGAYP-----
MLBr_01556|M.leprae_Br4923 GEMVTATQSVGDETLELLGSEMNYNVQVVSPEDEDRELLEPFDLTYGEDQ
.
MUL_0889|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1128|M.marinum_M --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 GDPNGGGGGAGGAGGKGGDGGAQAGDGGAGGAGGKGGNGGNGATGATGLN
Rv1452c|M.tuberculosis_H37Rv ----------------------------SGAVGGKGNSGGN---------
Mflv_5091|M.gilvum_PYR-GCK G--------------------------GTGGTGGQGGTGGTGGTG-----
Mvan_1264|M.vanbaalenii_PYR-1 G--------------------------GTGGTGGQGGTGGTGGTG-----
TH_3810|M.thermoresistible__bu --------------------------------------------------
MSMEG_3545|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3220|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2780|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 GDEDELQVRPPVVTVMGHVDHGKTRLLDTIRKANVREAEAGGITQHIG--
MUL_0889|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1128|M.marinum_M --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 GLGAGADGTDGGKGGNGGAGGGGGAGGQGGKALAATHQDGSMGAGGAGGN
Rv1452c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_5091|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1264|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MSMEG_3545|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3220|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2780|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MUL_0889|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1128|M.marinum_M --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 GGAGGMGGDGGNGAKGTFDNGGDGVGGNGGNGGSRGIGGAGGIGGAGSTA
Rv1452c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_5091|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1264|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MSMEG_3545|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3220|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2780|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MUL_0889|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1128|M.marinum_M --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 GADGARGATPTSGGNGGTGGNGANATVAGGAGGAGGKGGNGGLVGNGGAG
Rv1452c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_5091|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1264|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MSMEG_3545|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3220|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2780|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MUL_0889|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1128|M.marinum_M --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 GKGGDGMAGVAGSSPTTAGESGTSGQNGGAGGAGGAGGRGGDFGGDGGTG
Rv1452c|M.tuberculosis_H37Rv ------------------------------GGQGGAGGDGGTLAGNGGAG
Mflv_5091|M.gilvum_PYR-GCK ----------------------------GTGGTGGAGGDGG-LGGLGGFG
Mvan_1264|M.vanbaalenii_PYR-1 ----------------------------GTGGTGGAGGDGG-LGGLGGFG
TH_3810|M.thermoresistible__bu -------------------------------------------SGNGVFS
MSMEG_3545|M.smegmatis_MC2_155 --------------------------------------------NRHDWW
MAB_3220|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2780|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 -----------------------AYQVGVDLDGSERLITFIDTPGHEAFT
MUL_0889|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1128|M.marinum_M --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 GAGGNGADGGAGGNGANGANATTPGAKGGDGGHGGPGAQGGNGGQGGPGG
Rv1452c|M.tuberculosis_H37Rv GTGGRGADGGLGGSGAEGANATTAGERGQDGGKGG------NGG------
Mflv_5091|M.gilvum_PYR-GCK GSGGTGGQGGLGGPGGSTGGAGGAGGQAGTAGKGGLFGGSGSAG------
Mvan_1264|M.vanbaalenii_PYR-1 GVGGTGGQGGQGGPGGSTGGAGGAGGQAGVAGKGGLFGGSGSAG------
TH_3810|M.thermoresistible__bu GGGGTPSAG-FGVPTTIQFPAGSCTSRAVSR-------------------
MSMEG_3545|M.smegmatis_MC2_155 DDRQGPPPWGWGPPPPVHWNGGPLPHTINYWG------------------
MAB_3220|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2780|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 AMRARGAKATDIAILVVAADDGVMPQTVEAINHAQAADVPIVVAVNKIDK
MUL_0889|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1128|M.marinum_M --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 LAGNLFGQNGIQGVGGSGGKGGAGGLAGDGGNGANGNFAFGDGNGGHGGN
Rv1452c|M.tuberculosis_H37Rv -------------VGGTGGNAVAP--------GAN---------GGHGGN
Mflv_5091|M.gilvum_PYR-GCK ------------KAGATGAAGASGGKGAQGATGAQG-------ATGAQGA
Mvan_1264|M.vanbaalenii_PYR-1 ------------TAGVTGASGASGGKGAQGATGAQG-------ATGAQGA
TH_3810|M.thermoresistible__bu ----------------------------------------------EGIM
MSMEG_3545|M.smegmatis_MC2_155 -----------------------------------------------YNL
MAB_3220|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2780|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 EGADPAKIRGQLTEYGLVAEDFGGDTMFIDISAKVGTNIEALLEAVLLTA
MUL_0889|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1128|M.marinum_M --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 GGNPGAGGQGGSGGAGSTPGAKGAHGFTPTSGGDGGDGGNGGHSQVVGGN
Rv1452c|M.tuberculosis_H37Rv GGNPGFSGAGGLGGLSGDGVTRAAQGATPDFADTGGKGGNGGN-------
Mflv_5091|M.gilvum_PYR-GCK TGATGAAGKAGATGAAGAAGAAGSAGTTGAQGSAGTNGANGAN-------
Mvan_1264|M.vanbaalenii_PYR-1 TGATGASGAQGSAGSAGSAGVAGSAGTTGAQGSAGTNGANGAN-------
TH_3810|M.thermoresistible__bu PSSRAASARAGAACASSTSRSRRSFCCSRA--------------------
MSMEG_3545|M.smegmatis_MC2_155 NPVWHDGFRQWGVWLFGIWIPIFGVGFN----------------------
MAB_3220|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2780|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 DAALDLRANSGMEAQGVAIEAHLDRGRGPVATVLVQRGTLRIGDSVVAGD
MUL_0889|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1128|M.marinum_M --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 GGDGGNGGNGGSAGTGGNGGRGGDGAFGGMSANATNPGENGPNGNPGGNG
Rv1452c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_5091|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1264|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MSMEG_3545|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3220|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2780|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 AYGRVRRMVDEHGVDIEAALPSSPVQVIGFTSVPGAGDNFLVVDEDRIAR
MUL_0889|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1128|M.marinum_M --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 GAGGAGGAGLNGGNGGAGGNGGLGGFGGNGAAGANGVAVGAPGQPGGAGG
Rv1452c|M.tuberculosis_H37Rv --------------------------------GANAVAP------GGTGA
Mflv_5091|M.gilvum_PYR-GCK --------------------------------GSQGAAG----KAGATGT
Mvan_1264|M.vanbaalenii_PYR-1 --------------------------------GSQGAAG----KAGATGT
TH_3810|M.thermoresistible__bu --------------------------------------------------
MSMEG_3545|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3220|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2780|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 QIADRRSARKRNALAARSRKRISLEDLDSALKETSQLNLILKGDNAGTVE
MUL_0889|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1128|M.marinum_M --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 HGGAGGNGGAGGNGGQG-VVSDGAGGAGGAGGDGGAPGDGANGGNGQGAG
Rv1452c|M.tuberculosis_H37Rv SGGAGGNAGAGGKGGEN-IIGDGGGGNGGAGGKG-------------GAG
Mflv_5091|M.gilvum_PYR-GCK TGATGAAGEDGDAGSGGSGINGGAGGAGGAGGEG-------------GEG
Mvan_1264|M.vanbaalenii_PYR-1 TGATGAAGDAGDPGSEAVDINGGAGGAGGAGGEG-------------GEG
TH_3810|M.thermoresistible__bu --------------------------------------------------
MSMEG_3545|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3220|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2780|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 ALEEALMGIQVDDEVALRVIDRGVGGITETNVNLASASDAIIIGFNVRAE
MUL_0889|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1128|M.marinum_M --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 AFAGGGGGRGGDGGNAGNAGAGGPGGTGSTAGKAGPAGSILHDGGNGGHG
Rv1452c|M.tuberculosis_H37Rv TLLG----------------------------------------------
Mflv_5091|M.gilvum_PYR-GCK GDGG----------------------------------------------
Mvan_1264|M.vanbaalenii_PYR-1 GAGG----------------------------------------------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MSMEG_3545|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3220|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2780|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 GKATELASREG---------------------------------------
MUL_0889|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1128|M.marinum_M --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 GHGAASGGNGGPGGHGGNGGNGGTGANGGNGGIGGTGGAGSTGAKGVLGT
Rv1452c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_5091|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1264|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MSMEG_3545|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3220|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2780|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MUL_0889|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1128|M.marinum_M --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 NEGDGGDGGRGGNGGRGGNGGQGLTGAGGNGGTGGTPGNGGNGGNGASGD
Rv1452c|M.tuberculosis_H37Rv -----------------------LTVFGDNGGAG----------------
Mflv_5091|M.gilvum_PYR-GCK --------------------------KGGAGAEG----------------
Mvan_1264|M.vanbaalenii_PYR-1 --------------------------AGGDGSEG----------------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MSMEG_3545|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3220|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2780|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MUL_0889|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1128|M.marinum_M --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 LVTSPGDGGGGGRGGDAGRGGDAGLGGSSGPGGTPGDWGTGGTGGTGGTG
Rv1452c|M.tuberculosis_H37Rv -----------------------VLGDSTDPDGS----------------
Mflv_5091|M.gilvum_PYR-GCK -----------------------GAGGGGGAGGS----------------
Mvan_1264|M.vanbaalenii_PYR-1 -----------------------GAGGSGGAGGS----------------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MSMEG_3545|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3220|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2780|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 ------------VEIRYYLVIYQAIDDIEKALRGMLKPIYEENQLGRAEI
MUL_0889|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1128|M.marinum_M --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 GQGANGGLTGGRGGTGGNGGAGGTGGTGHNGSQPGMGGNGGAGGFGGNGF
Rv1452c|M.tuberculosis_H37Rv ------------GGAGGAGGAGGAGGD------PTI--------------
Mflv_5091|M.gilvum_PYR-GCK -------------GAGGAGGAGGSGGS-----------------------
Mvan_1264|M.vanbaalenii_PYR-1 -------------GTGGAGGAGGSGGS-----------------------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MSMEG_3545|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3220|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2780|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 RALFRSSKVGIIAGCIISSGVVRRNAKVRLLRDNIVVTDNLTVTSLRREK
MUL_0889|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1128|M.marinum_M --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 AGVGGRGGMGGSGGTGGTGDAGPFGTGTGGTGGHGGQGGGGGFSILLGLG
Rv1452c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_5091|M.gilvum_PYR-GCK ---GGSGGSGGSGGAAGDSDD-----------------------------
Mvan_1264|M.vanbaalenii_PYR-1 ---GGSGGAGGSGGAAGDSD------------------------------
TH_3810|M.thermoresistible__bu --------------------------------------------------
MSMEG_3545|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3220|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2780|M.avium_104 --------------------------------------------------
MLBr_01556|M.leprae_Br4923 DDVTEVREGFECGMTLGYSDIKEGDVIESYELVEKQRA------------
MUL_0889|M.ulcerans_Agy99 ----------------------------------
MMAR_1128|M.marinum_M ----------------------------------
Mb1485c|M.bovis_AF2122/97 GLGGLGSPGSIATGTAGGAGGGGGFGGLGGGEFV
Rv1452c|M.tuberculosis_H37Rv ----------------------------------
Mflv_5091|M.gilvum_PYR-GCK ----------------------------------
Mvan_1264|M.vanbaalenii_PYR-1 ----------------------------------
TH_3810|M.thermoresistible__bu ----------------------------------
MSMEG_3545|M.smegmatis_MC2_155 ----------------------------------
MAB_3220|M.abscessus_ATCC_1997 ----------------------------------
MAV_2780|M.avium_104 ----------------------------------
MLBr_01556|M.leprae_Br4923 ----------------------------------