For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTSGSVALRHVRQQVRPDVSLHAVIAGSGPAVFLLHGWPQTWQEWLPILEDLARHHTVVAVDLKGAGGS SKPLLGYDKVTMAEELDILREKLGFDTVQVVGHDIGGMLAYAWAATHRDTVTRLAVFDVPIPGASLWDGI LTDPLAWHFAFHQCRDVPEFLIAGRERGYVEYFLRGRTANQDAFTDAAVDHYAHALAQPGAVRSTLEWYR AFAQDADDNRKLAREPLTVPVLALGGEHRWGPKMVDVLREFATDVRGGSIPDCGHWVVEERPDVVLAELE AFLQR
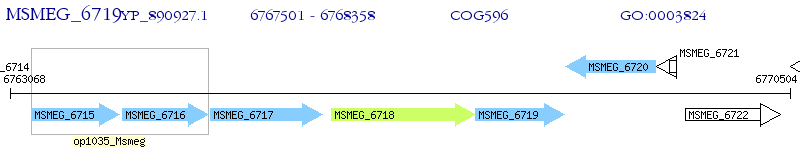
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6719 | - | - | 100% (285) | epoxide hydrolase |
| M. smegmatis MC2 155 | MSMEG_6720 | - | 7e-51 | 40.22% (271) | epoxide hydrolase |
| M. smegmatis MC2 155 | MSMEG_6708 | - | 5e-37 | 36.62% (284) | epoxide hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3694 | ephE | 2e-19 | 31.75% (274) | epoxide hydrolase EphE |
| M. gilvum PYR-GCK | Mflv_1377 | - | 2e-20 | 30.86% (269) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | Rv0134 | ephF | 3e-27 | 31.93% (285) | epoxide hydrolase EphF |
| M. leprae Br4923 | MLBr_02297 | - | 4e-23 | 32.97% (276) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_4522c | - | 8e-22 | 28.26% (276) | epoxide hydrolase EphF |
| M. marinum M | MMAR_0342 | ephF | 2e-24 | 30.07% (276) | epoxide hydrolase EphF |
| M. avium 104 | MAV_5163 | - | 1e-26 | 31.54% (279) | hydrolase, alpha/beta fold family protein |
| M. thermoresistible (build 8) | TH_4292 | - | 2e-41 | 35.40% (274) | PUTATIVE putative epoxide hydrolase |
| M. ulcerans Agy99 | MUL_4778 | ephF | 2e-24 | 30.07% (276) | epoxide hydrolase EphF |
| M. vanbaalenii PYR-1 | Mvan_5429 | - | 4e-21 | 33.70% (270) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6719|M.smegmatis_MC2_155 --MTTSGSVALRHVRQQVR-------------PDVSLHAVIAG-------
TH_4292|M.thermoresistible__bu --VVDTGAHAFDSATRDDDEFNRLFRHEFADVDGVRMHYVTGG-------
Rv0134|M.tuberculosis_H37Rv ----MIALPALEGVEHRHVDVAE----------GVRIHVADAGPA-----
MAV_5163|M.avium_104 ----MVTMPELDGVEHRYVDLGD----------DVTIHVADAGPA-----
MMAR_0342|M.marinum_M ----MSTLPALDGVEHRFVDLDG----------GITIHVADAGPA-----
MUL_4778|M.ulcerans_Agy99 ----MSTLPALDGVEHRFVDLDG----------GITIHVADAGPA-----
MAB_4522c|M.abscessus_ATCC_199 --MTQRSMPVLDGVEHQYVQAG-----------DVKIHVAVAGPQ-----
Mb3694|M.bovis_AF2122/97 MAAPDPSMTRIAGPWRHLDVHAN----------GIRFHVVEAVPSGQPEG
MLBr_02297|M.leprae_Br4923 MAMSNPSVTRIAGPWHHLDVHAN----------GIRFHVVEAVPT-QPAG
Mflv_1377|M.gilvum_PYR-GCK MTPPDPSVVRIGGPWRHLDVHAN----------GIRFHVVEAQRP---SG
Mvan_5429|M.vanbaalenii_PYR-1 MVRPDPSVVRIGGPWRHLDVHAN----------GIRFHVVEAERQ---AG
: : .: :* . .
MSMEG_6719|M.smegmatis_MC2_155 ------------SGPAVFLLHGWPQTWQEWLPILEDLARH-HTVVAVDLK
TH_4292|M.thermoresistible__bu ------------SGDPLVLLHGWPQTWYAWRGIIPALAQK-YTVYALDLP
Rv0134|M.tuberculosis_H37Rv ------------DGPAVMLVHGFPQNWWEWRDLIGPLAADGNRVLCPDLR
MAV_5163|M.avium_104 ------------SGPAVMLVHGFPQNWWEWRALIGPLAADGYRVLCPDLR
MMAR_0342|M.marinum_M ------------DGPAVMLVHGFPENWWEWHELIGPLAADGYRVLCPDLR
MUL_4778|M.ulcerans_Agy99 ------------DGPAVMLVHGFPENWWEWHELIGPLAADGYRVLCPDLR
MAB_4522c|M.abscessus_ATCC_199 ------------DGRAVVLLHGWPENWWMWHKVIPALAAAGYRVHAMDLR
Mb3694|M.bovis_AF2122/97 PDAATPPMQPALARPLVILLHGFGSFWWSWRHQLCGLTGA--RVVAVDLR
MLBr_02297|M.leprae_Br4923 QDCAP----SATARPLVMLLHGFGSFWWSWRHQLRGLTEA--RLVAVDLR
Mflv_1377|M.gilvum_PYR-GCK ADDVT---RPLTDRPLVILLHGFGSFWWSWRHQLTGLSGA--RLVAVDLR
Mvan_5429|M.vanbaalenii_PYR-1 ADDRS---KPLTDRPLVILLHGFGSFWWSWRHQLAGLTGA--RVVAVDLR
:.*:**: . * * : *: : . **
MSMEG_6719|M.smegmatis_MC2_155 GAGGSSKPLLGYDKVTMAEELD-ILREKLGFDTVQVVGHDIGGMLAYAWA
TH_4292|M.thermoresistible__bu GLGDSEGAPPSYDKATLARFVHRLLADQLGLRSIRLVAHDLGAGVGFQYA
Rv0134|M.tuberculosis_H37Rv GAGWSSAPRSRYTKTEMADDLA-AVLDGLGVAKVKLVAHDWGGPVAFIMM
MAV_5163|M.avium_104 GAGWSSAPRSSYRKDEMADDLA-GVLDRLGVRSVDLVAHDWGGPVAFIMM
MMAR_0342|M.marinum_M GAGWSSAPRSRYTKKEMADDLA-GVLDRLGIASVRLVAHDWGGPVAFIMM
MUL_4778|M.ulcerans_Agy99 GAGWSSAPRSRYTKKEMADDLA-GVLDRLGIASVRLVAHDWGGPVAFIMM
MAB_4522c|M.abscessus_ATCC_199 GAGWSEVAPAGYEKEQFASDVL-AAADALGLESFDLVGHDWGGWTAQLVA
Mb3694|M.bovis_AF2122/97 GYGGSDKPPRGYDGWTLAGDTA-GLIRALGHPSATLVGHADGGLACWTTA
MLBr_02297|M.leprae_Br4923 GYGGSDKPPRGYDGWTLAGDTA-GLIRALGHSSATLVGHAEGGLACWATA
Mflv_1377|M.gilvum_PYR-GCK GYGGSDKPPRGYDGWTLAGDTA-GLVRALGHNSATLVGHADGGLVCWATS
Mvan_5429|M.vanbaalenii_PYR-1 GYGGSDKPPRGYDGWTLAGDAA-GLVRALGHNTATLVGHADGGLVCWATS
* * *. . * :* ** . :*.* *.
MSMEG_6719|M.smegmatis_MC2_155 ATHRDTVTRLAVFDVPIPGASLWDGILTD-------PLAWHFAFHQCRDV
TH_4292|M.thermoresistible__bu AQYPTEVSKYAHLDYPLPGPALSAARYR--------EFSWHMAFHHQPRV
Rv0134|M.tuberculosis_H37Rv LRHPEKVTGFFGVNTVAPWVKRDLGMLRN-------MWRFWYQIPMSLPV
MAV_5163|M.avium_104 LRHPEKVTGFFGVNTSAPFVKRSLSTIRN-------VWRFWYQIPISLPV
MMAR_0342|M.marinum_M LRYPAKVTGFFGMNTAAPWFPRDLGMVRD-------IWRFWYQIPIMLPV
MUL_4778|M.ulcerans_Agy99 LRYPAKVTGFFGMNTAAPWFPRDLGMVRD-------IWRFWYQIPILLPV
MAB_4522c|M.abscessus_ATCC_199 LKAQSRIRRLVVLNIPPVWQQPGR-VARY-------AHKLAYQLVVGAPV
Mb3694|M.bovis_AF2122/97 LLHSRLVRAIALISSPHPAALRRSTLTRRDQRHALLPTLLRYQLPIWPER
MLBr_02297|M.leprae_Br4923 LLHPRLVRAIALINSPHPAALRRSMLTHRNQGYALLPTLLRYQLPIWPER
Mflv_1377|M.gilvum_PYR-GCK VLHPRVVKAIAVVSSPHPSALRASALRRRDQGRALLPSMMRYQVPMWPER
Mvan_5429|M.vanbaalenii_PYR-1 VLHPRVVQAIAVVSSPHPAALRASALTRRDQGQALLPSLLRYQVPIRPER
: .. .
MSMEG_6719|M.smegmatis_MC2_155 PEFLIAGRERGYVEYFLR---------GRTANQDAFTDAAVDHYAHALAQ
TH_4292|M.thermoresistible__bu PEAVVANDVREYLALFYPYVAYGGTSFGGPGAQSPFTDEQVDEFARTYNR
Rv0134|M.tuberculosis_H37Rv IGPRVISDPKGRYFRLLTG---------WVGGGFRVPDDDVRLYLDCMRE
MAV_5163|M.avium_104 IGPRVISTPDSRFVRLLGS---------WVGGGYTLPEEDVRLYLECMRQ
MMAR_0342|M.marinum_M IGPRVIADPKARYFRVLTS---------WVGGGFTIADEDVRMYVERMRQ
MUL_4778|M.ulcerans_Agy99 IGPRVIADPKARYFRVLTS---------WVGGGFTIADEDVRMYVERMRQ
MAB_4522c|M.abscessus_ATCC_199 VGPLSHLSPTLWWFIRRS------------G----MPKESVDFFRSCFRD
Mb3694|M.bovis_AF2122/97 LLTRNNAAEIERLVRARGC---------AKWLASEDFSQAIDHLRQAIQI
MLBr_02297|M.leprae_Br4923 LLTRNNAAEIERLVRSRSS---------AKWQACEDFAVTIDHLRIAIQI
Mflv_1377|M.gilvum_PYR-GCK ALTRHNGAELERLVRSRAS---------EKWQVSEDFAETVGHLRRAVQI
Mvan_5429|M.vanbaalenii_PYR-1 LLTRDDGAELERLVRSRSS---------AKWQAGEDFSESIRHMRRAIQI
:
MSMEG_6719|M.smegmatis_MC2_155 PGAVRSTLEWYRAFAQD-----ADDNRKLAREPLTVPVLALGG--EHRWG
TH_4292|M.thermoresistible__bu PDVLSGGFDLYRTLDQD-----EHDN--VAAGPITTPTLLMTA--EGLLE
Rv0134|M.tuberculosis_H37Rv PGHAEAGSRWYRTFQTREMLRWLRGEYNDARVDVPVRWLHGTG--DPVIT
MAV_5163|M.avium_104 PGHAEAGSRWYRSFQTREMLSWMRGEYDGARVHVPVRWLSGTE--DPVLT
MMAR_0342|M.marinum_M PGHAVAGSRWYRTFQSSEALPWMRGEYADARVDVPVRWLHGTE--DPVLT
MUL_4778|M.ulcerans_Agy99 PGHAVAGSRWYRTFQSSEALPWMRGEYADARVDVPVRWLHGTD--DPVLT
MAB_4522c|M.abscessus_ATCC_199 RDRRVAGSKIYRSMVGKEFANLARGPYDDQKLTMPVRVLFGLD--DVAVH
Mb3694|M.bovis_AF2122/97 PAAAHCALEYQRWAVRSQLRSEGRRFIRAMTQQLGMPLLHLRGDADPYVL
MLBr_02297|M.leprae_Br4923 PAAAHCALEYQRWAVRSQLRNEGRKFMKSMSRPFSVPLLHVRGDADPYLL
Mflv_1377|M.gilvum_PYR-GCK PSAAHCALEYQRWAVRSQLRAEGRRFMRSMRAPLSVPMLHLRGDADPYVL
Mvan_5429|M.vanbaalenii_PYR-1 PSAAHCALEYQRWAVRSQLRAEGRRFMSSMKRPLGVPVLHLRGEQDPYVL
* . * :
MSMEG_6719|M.smegmatis_MC2_155 PKMVDVLREFATDVRGGSIPDCGHWVVEERPDVVLAELEAFLQR------
TH_4292|M.thermoresistible__bu STRLTVEPRMANITRAVEVPGAGHWLVEENPAFVTNELLTFLAD------
Rv0134|M.tuberculosis_H37Rv PDLLDGYAERASDFEVELVDGVGHWIVEQRPELVLDRVRAFLAAGTEQRD
MAV_5163|M.avium_104 PDLLDGYAERIDDFQLELVDGVGHWIVDQRPDLVLDRVRAFLRRET----
MMAR_0342|M.marinum_M PQLLRGYEDHISDFEVEFVDGVGHWIVEQRPDLVLDRLRAFLRIET----
MUL_4778|M.ulcerans_Agy99 PQLLRGYEDHISDFEVEFVDGVGHWIVEQRPDLVLDRLRAFLRIET----
MAB_4522c|M.abscessus_ATCC_199 PSLLDGFKDHADDLNITEVPNCGHFIVDEQPELVVKWLSEVLAEPAP---
Mb3694|M.bovis_AF2122/97 ADPVERTQRYAPHGRYISIAGAGHFSHEEAPEEVNRHLMRFLEQVH--QL
MLBr_02297|M.leprae_Br4923 ADSVDRTRRYAPHGRYVSVSDAGHFSHEEAPEEINRYLTRFLKQVHGPRL
Mflv_1377|M.gilvum_PYR-GCK PDPVYRTQRYAPHGRYVSIAGAGHFAHEEQPAAVNAQLSRFLSQVYT---
Mvan_5429|M.vanbaalenii_PYR-1 ADPVYRTQRYAPHGRYVSIEGAGHFSHEEAPAAVNEQLRRFLNQVYG---
. : . : . **: :: * : : .*
MSMEG_6719|M.smegmatis_MC2_155 -
TH_4292|M.thermoresistible__bu -
Rv0134|M.tuberculosis_H37Rv -
MAV_5163|M.avium_104 -
MMAR_0342|M.marinum_M -
MUL_4778|M.ulcerans_Agy99 -
MAB_4522c|M.abscessus_ATCC_199 -
Mb3694|M.bovis_AF2122/97 S
MLBr_02297|M.leprae_Br4923 S
Mflv_1377|M.gilvum_PYR-GCK -
Mvan_5429|M.vanbaalenii_PYR-1 -