For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRLAGKIALITGAANGMGCATAETFSRQGATVVIADVDDEDGLQVAKQIGTSGGTAHFVHLDVTDETAWA CAVEDVLGRYERLDILVNNAGISGTFDPDLTSTAFFDRLIAVNARGVFLGIKHGAAAMKHTGGGSIVNLS SISAHIGQLGVHLGYGASKAAVKAMTTTAAVHYAADGIRVNAVAPGMLPPMQTSRGSADPAWRAKQIDGV PLKREGHVQEVADAVLFLASDESSYITGTELMVDGGLTAV
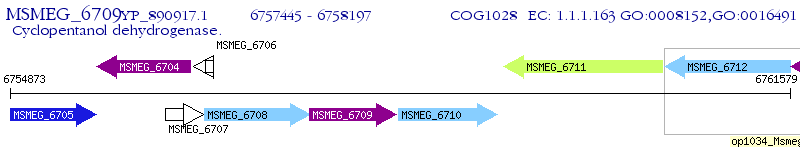
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6709 | - | - | 100% (250) | cyclopentanol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2477 | - | 2e-45 | 43.32% (247) | cyclopentanol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3515 | - | 3e-43 | 42.28% (246) | 3-alpha-(or 20-beta)-hydroxysteroid dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2025 | fabG3 | 2e-43 | 39.92% (248) | 20-beta-hydroxysteroid dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4505 | - | 1e-44 | 43.78% (249) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2002 | fabG3 | 2e-43 | 39.92% (248) | 20-beta-hydroxysteroid dehydrogenase |
| M. leprae Br4923 | MLBr_00862 | ephD | 6e-21 | 33.19% (226) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4680c | - | 9e-41 | 40.73% (248) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_2981 | fabG3_1 | 2e-44 | 39.11% (248) | 20-beta-hydroxysteroid dehydrogenase FabG3_1 |
| M. avium 104 | MAV_2509 | - | 2e-46 | 40.73% (248) | 3-alpha-(or 20-beta)-hydroxysteroid dehydrogenase |
| M. thermoresistible (build 8) | TH_4254 | - | 3e-48 | 39.52% (248) | PUTATIVE putative 2,5-dichloro-2,5-cyclohexadiene-1,4-diol |
| M. ulcerans Agy99 | MUL_1037 | fabG3 | 4e-42 | 40.24% (246) | 20-beta-hydroxysteroid dehydrogenase FabG3 |
| M. vanbaalenii PYR-1 | Mvan_3000 | - | 2e-43 | 42.34% (248) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2025|M.bovis_AF2122/97 --------------------------------------------------
Rv2002|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2981|M.marinum_M --------------------------------------------------
MAV_2509|M.avium_104 --------------------------------------------------
Mflv_4505|M.gilvum_PYR-GCK --------------------------------------------------
MUL_1037|M.ulcerans_Agy99 --------------------------------------------------
Mvan_3000|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4680c|M.abscessus_ATCC_199 --------------------------------------------------
TH_4254|M.thermoresistible__bu --------------------------------------------------
MSMEG_6709|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIVLVHGFPDSHVLWD
Mb2025|M.bovis_AF2122/97 --------------------------------------------------
Rv2002|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2981|M.marinum_M --------------------------------------------------
MAV_2509|M.avium_104 --------------------------------------------------
Mflv_4505|M.gilvum_PYR-GCK --------------------------------------------------
MUL_1037|M.ulcerans_Agy99 --------------------------------------------------
Mvan_3000|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4680c|M.abscessus_ATCC_199 --------------------------------------------------
TH_4254|M.thermoresistible__bu --------------------------------------------------
MSMEG_6709|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVVPLLAKRFRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDELSP
Mb2025|M.bovis_AF2122/97 --------------------------------------------------
Rv2002|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2981|M.marinum_M --------------------------------------------------
MAV_2509|M.avium_104 --------------------------------------------------
Mflv_4505|M.gilvum_PYR-GCK --------------------------------------------------
MUL_1037|M.ulcerans_Agy99 --------------------------------------------------
Mvan_3000|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4680c|M.abscessus_ATCC_199 --------------------------------------------------
TH_4254|M.thermoresistible__bu --------------------------------------------------
MSMEG_6709|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSG
Mb2025|M.bovis_AF2122/97 --------------------------------------------------
Rv2002|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2981|M.marinum_M --------------------------------------------------
MAV_2509|M.avium_104 --------------------------------------------------
Mflv_4505|M.gilvum_PYR-GCK --------------------------------------------------
MUL_1037|M.ulcerans_Agy99 --------------------------------------------------
Mvan_3000|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4680c|M.abscessus_ATCC_199 --------------------------------------------------
TH_4254|M.thermoresistible__bu --------------------------------------------------
MSMEG_6709|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMV
Mb2025|M.bovis_AF2122/97 --------------------------------------------------
Rv2002|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2981|M.marinum_M --------------------------------------------------
MAV_2509|M.avium_104 --------------------------------------------------
Mflv_4505|M.gilvum_PYR-GCK --------------------------------------------------
MUL_1037|M.ulcerans_Agy99 --------------------------------------------------
Mvan_3000|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4680c|M.abscessus_ATCC_199 --------------------------------------------------
TH_4254|M.thermoresistible__bu --------------------------------------------------
MSMEG_6709|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQ
Mb2025|M.bovis_AF2122/97 --------------------------------------------------
Rv2002|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2981|M.marinum_M --------------------------------------------------
MAV_2509|M.avium_104 --------------------------------------------------
Mflv_4505|M.gilvum_PYR-GCK --------------------------------------------------
MUL_1037|M.ulcerans_Agy99 --------------------------------------------------
Mvan_3000|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4680c|M.abscessus_ATCC_199 --------------------------------------------------
TH_4254|M.thermoresistible__bu --------------------------------------------------
MSMEG_6709|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAAVHD
Mb2025|M.bovis_AF2122/97 ------------------MSGR---LIGKVALVSGGARGMGASHVRAMVA
Rv2002|M.tuberculosis_H37Rv ------------------MSGR---LIGKVALVSGGARGMGASHVRAMVA
MMAR_2981|M.marinum_M ------------------MAGR---LTGKVALVSGGARGMGASHVRALVA
MAV_2509|M.avium_104 ------------------MAER---LAGKVALVSGGARGMGASHVRSLVA
Mflv_4505|M.gilvum_PYR-GCK ------------------MTGR---LAGKVALISGAARGMGASHARVMAG
MUL_1037|M.ulcerans_Agy99 -------------------MGR---VDGKVALISGGARGMGASHARLLVQ
Mvan_3000|M.vanbaalenii_PYR-1 -------------------MGR---VDGKVALISGGAQGMGAADARALIA
MAB_4680c|M.abscessus_ATCC_199 ------------------MNGNSALLQGKNVIVTGGARGLGAAFSRHIVS
TH_4254|M.thermoresistible__bu ------------------MTAGSTRLAGKVVLVTGAGGGLGTAIARRLAD
MSMEG_6709|M.smegmatis_MC2_155 --------------------MR---LAGKIALITGAANGMGCATAETFSR
MLBr_00862|M.leprae_Br4923 LADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAGSGIGRETALALAR
. .. . ::*.. *:* :
Mb2025|M.bovis_AF2122/97 EGAKVVFGDILDEEGKAVAAELA----DAARYVHLDVTQPAQWTAAVDTA
Rv2002|M.tuberculosis_H37Rv EGAKVVFGDILDEEGKAVAAELA----DAARYVHLDVTQPAQWTAAVDTA
MMAR_2981|M.marinum_M EGAHVVLGDILDDEGRAVAAELG----DAARYVHLDVTQPEQWTAAVDTA
MAV_2509|M.avium_104 EGAKVVFGDILDDEGKAVAAEVG----EATRYLHLDVTKPEDWDAAVATA
Mflv_4505|M.gilvum_PYR-GCK HGAKVVCGDILDSDGEAVAAELG----DAARYVHLDVTSPDDWDRAVAAA
MUL_1037|M.ulcerans_Agy99 EGAKVVIGDILDEEGKALAEEIG----DAARYVHLDVTQPDQWEAAVATA
Mvan_3000|M.vanbaalenii_PYR-1 EGAKVVIGDILDDKGKALADEINAETPDSIRYVHLDVTQADQWEAAVATA
MAB_4680c|M.abscessus_ATCC_199 QGGRVVIADVLDGDGRQLAAELG----DAARFTHLDVTDAEQWRELIDTT
TH_4254|M.thermoresistible__bu EGARLVLADLQESQCAALADELPAP-AGAHHPLGFDVADETAWISAISTV
MSMEG_6709|M.smegmatis_MC2_155 QGATVVIADVDDEDGLQVAKQIGTS-GGTAHFVHLDVTDETAWACAVEDV
MLBr_00862|M.leprae_Br4923 RGAEVVVSDIDEAAAKNTAAQIVAS-GGVAYAYALDVSDAAAVEAFAEQV
.*. :* .*: : * :: :**:. .
Mb2025|M.bovis_AF2122/97 VTAFGGLHVLVNNAGILNIGTIEDYALTEWQRILDVNLTGVFLGIRAVVK
Rv2002|M.tuberculosis_H37Rv VTAFGGLHVLVNNAGILNIGTIEDYALTEWQRILDVNLTGVFLGIRAVVK
MMAR_2981|M.marinum_M VNEFGGLHVLVNNAGILNIGTIEDYALSEWQRILDINVTGVFLGIRAAVK
MAV_2509|M.avium_104 LAEFGRIDVLVNNAGIINIGTLEDYALSEWQRILDINLTGVFLGIRAVVK
Mflv_4505|M.gilvum_PYR-GCK VADFGGLDVLVNNAGILNIGTVEDYELSEWHRILDVNLTGVFLGIRAATP
MUL_1037|M.ulcerans_Agy99 VDEFGKLDVLVNNVGIVALGQLKKFDLGKWQKVIDVNLTGTFLGMRAAVE
Mvan_3000|M.vanbaalenii_PYR-1 VDAFGKLNVLVNNAGTVALGQIGQFDMAKWQKVIDVNLTGTFLGMQASVE
MAB_4680c|M.abscessus_ATCC_199 LAGFGTITGLVNNAGISSAAMVADEPLEHFRKVIEINLVGVFIGMQAVIP
TH_4254|M.thermoresistible__bu SAQYGALHALVNNAAIGALGNVAEERLERWDRILDVGATGVWLGMKHAVP
MSMEG_6709|M.smegmatis_MC2_155 LGRYERLDILVNNAGISGTFDPDLTSTAFFDRLIAVNARGVFLGIKHGAA
MLBr_00862|M.leprae_Br4923 SAKHGVPDIVVNNAGIGQAGRFLDTPPEEFDRVLAVNLGGVVNCCRAFGQ
. :***.. : ::: :. *. :
Mb2025|M.bovis_AF2122/97 PMKEAGRG-SIINISSIEGLAG-TVACHGYTATKFAVRGLTKSTALELGP
Rv2002|M.tuberculosis_H37Rv PMKEAGRG-SIINISSIEGLAG-TVACHGYTATKFAVRGLTKSTALELGP
MMAR_2981|M.marinum_M PMKEAGRG-SIINISSIEGLAG-TIASHGYTVSKFAVRGLTKSTALELGP
MAV_2509|M.avium_104 PMKKAGRG-SIINISSIEGMAG-TIACHGYTATKFAVRGLTKSAALELGP
Mflv_4505|M.gilvum_PYR-GCK TMKAAGRG-SIINIASIEGMAG-TVGCHGYTATKFAVRGLTKSTALELGP
MUL_1037|M.ulcerans_Agy99 PMTAAGSG-SIINVSSIEGLRG-APAVHPYVASKWAVRGLTKSAALELAP
Mvan_3000|M.vanbaalenii_PYR-1 AMKTAGGG-SIINISSIEGLRG-AVMVHPYVASKWAVRGLTKSAALELGS
MAB_4680c|M.abscessus_ATCC_199 AMRAHGCG-SIVNISSAAGLVG-LALTSGYGASKWGVRGLSKVAAIELGR
TH_4254|M.thermoresistible__bu LIRRSGGG-SVVNICSIFGTVGGFGDSAAYHAAKGAVRTLTKNAALHWAP
MSMEG_6709|M.smegmatis_MC2_155 AMKHTGGG-SIVNLSSISAHIGQLGVHLGYGASKAAVKAMTTTAAVHYAA
MLBr_00862|M.leprae_Br4923 RLVERGTGGHIVNVSSMAAYAP-LQSLSAYCTSKAATYMFSDCLRAELDA
: * * ::*:.* . * .:* .. :: .
Mb2025|M.bovis_AF2122/97 SGIRVNSIHPGLVKTPMTDWV-------PEDIFQTALG---------RAA
Rv2002|M.tuberculosis_H37Rv SGIRVNSIHPGLVKTPMTDWV-------PEDIFQTALG---------RAA
MMAR_2981|M.marinum_M SGIRVNSIHPGLVKTPMTEWV-------PEDLFQTALG---------RAA
MAV_2509|M.avium_104 SGIRVNSIHPGLIKTPMTEWV-------PEDIFQSALG---------RAA
Mflv_4505|M.gilvum_PYR-GCK FGIRVNSVHPGLVKTPMADWV-------PEDIFQSALG---------RIA
MUL_1037|M.ulcerans_Agy99 LNIRVNSIHPGFIRTPMTANL-------PDDMVTIPLG---------RPA
Mvan_3000|M.vanbaalenii_PYR-1 HNIRVNSVHPGFIRTPMTKHF-------PDNMLRIPLG---------RPG
MAB_4680c|M.abscessus_ATCC_199 ERIRVNSVHPGVIYTPMTAKEGF--REGEGNMPIAAMG---------RVG
TH_4254|M.thermoresistible__bu DGVRVNSLHPGFIETPQLRELYKGTERYERMLAGTPLG---------RLG
MSMEG_6709|M.smegmatis_MC2_155 DGIRVNAVAPGMLPPMQTSRGSADPAWRAKQIDGVPLK---------REG
MLBr_00862|M.leprae_Br4923 VGVGLTTICPGVIDTNIIQSTRIDTPTAKQERVRDRRGQLDMMFRLRRYG
: :.:: **.: . * .
Mb2025|M.bovis_AF2122/97 EPVEVSNLVVYLASDESSYSTGAEFVVDGGTVAGLAHNDFGAVEVSSQPE
Rv2002|M.tuberculosis_H37Rv EPVEVSNLVVYLASDESSYSTGAEFVVDGGTVAGLAHNDFGAVEVSSQPE
MMAR_2981|M.marinum_M EPMEVSNLVVYLASDESSYSTGAEFVVDGGTVAGLAHKDFSAVDVAAQPE
MAV_2509|M.avium_104 EPKEVSNLVVYLASDESSYSTGSEFVVDGGTTAGLGHKDFSNVETDAQPD
Mflv_4505|M.gilvum_PYR-GCK QPHEVSNLVVYLASDESSYSTGAEFVVDGGTLAGLAHKDFSAVDVDQQPE
MUL_1037|M.ulcerans_Agy99 ESREVSTFVVFLASDDASYATGSEFVMDGGLVTDVPHKQF----------
Mvan_3000|M.vanbaalenii_PYR-1 QPEEVATFVVFLASDESRYATGAEFVMDGGLTNDVPHK------------
MAB_4680c|M.abscessus_ATCC_199 NADEVAGAVAYLLSDAASYVTGAELAVDGGWTAGPTLTTITGQ-------
TH_4254|M.thermoresistible__bu RPDEIAAVVAFLVSDDASFITGSEVYADGGWTAR----------------
MSMEG_6709|M.smegmatis_MC2_155 HVQEVADAVLFLASDESSYITGTELMVDGGLTAV----------------
MLBr_00862|M.leprae_Br4923 PDKVADTILSAIKKNKPIRPVAPEAYALYGISRVLPQVLRSTARIRVI--
: : .: . ...* *
Mb2025|M.bovis_AF2122/97 WVT
Rv2002|M.tuberculosis_H37Rv WVT
MMAR_2981|M.marinum_M WVT
MAV_2509|M.avium_104 WVT
Mflv_4505|M.gilvum_PYR-GCK WIT
MUL_1037|M.ulcerans_Agy99 ---
Mvan_3000|M.vanbaalenii_PYR-1 ---
MAB_4680c|M.abscessus_ATCC_199 ---
TH_4254|M.thermoresistible__bu ---
MSMEG_6709|M.smegmatis_MC2_155 ---
MLBr_00862|M.leprae_Br4923 ---