For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TGRLAGKVALISGAARGMGASHARVMAGHGAKVVCGDILDSDGEAVAAELGDAARYVHLDVTSPDDWDRA VAAAVADFGGLDVLVNNAGILNIGTVEDYELSEWHRILDVNLTGVFLGIRAATPTMKAAGRGSIINIASI EGMAGTVGCHGYTATKFAVRGLTKSTALELGPFGIRVNSVHPGLVKTPMADWVPEDIFQSALGRIAQPHE VSNLVVYLASDESSYSTGAEFVVDGGTLAGLAHKDFSAVDVDQQPEWIT*
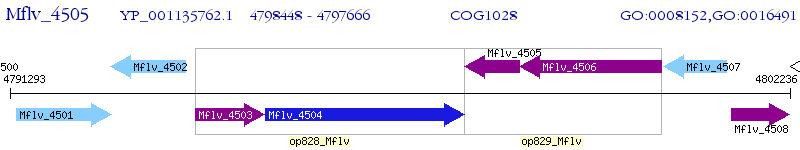
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4505 | - | - | 100% (260) | short-chain dehydrogenase/reductase SDR |
| M. gilvum PYR-GCK | Mflv_3281 | - | 3e-79 | 58.30% (247) | short-chain dehydrogenase/reductase SDR |
| M. gilvum PYR-GCK | Mflv_4369 | fabG | 3e-44 | 40.00% (240) | 3-ketoacyl-(acyl-carrier-protein) reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2025 | fabG3 | 1e-119 | 80.77% (260) | 20-beta-hydroxysteroid dehydrogenase |
| M. tuberculosis H37Rv | Rv2002 | fabG3 | 1e-119 | 80.77% (260) | 20-beta-hydroxysteroid dehydrogenase |
| M. leprae Br4923 | MLBr_01094 | - | 3e-25 | 35.57% (194) | short chain alcohol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4680c | - | 1e-57 | 46.12% (245) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_2981 | fabG3_1 | 1e-118 | 79.23% (260) | 20-beta-hydroxysteroid dehydrogenase FabG3_1 |
| M. avium 104 | MAV_2509 | - | 1e-118 | 78.60% (257) | 3-alpha-(or 20-beta)-hydroxysteroid dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3515 | - | 2e-84 | 61.32% (243) | 3-alpha-(or 20-beta)-hydroxysteroid dehydrogenase |
| M. thermoresistible (build 8) | TH_4254 | - | 2e-46 | 41.53% (248) | PUTATIVE putative 2,5-dichloro-2,5-cyclohexadiene-1,4-diol |
| M. ulcerans Agy99 | MUL_1037 | fabG3 | 8e-86 | 61.22% (245) | 20-beta-hydroxysteroid dehydrogenase FabG3 |
| M. vanbaalenii PYR-1 | Mvan_3000 | - | 3e-77 | 57.89% (247) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2025|M.bovis_AF2122/97 MSGR---LIGKVALVSGGARGMGASHVRAMVAEGAKVVFGDILDEEGKAV
Rv2002|M.tuberculosis_H37Rv MSGR---LIGKVALVSGGARGMGASHVRAMVAEGAKVVFGDILDEEGKAV
MMAR_2981|M.marinum_M MAGR---LTGKVALVSGGARGMGASHVRALVAEGAHVVLGDILDDEGRAV
MAV_2509|M.avium_104 MAER---LAGKVALVSGGARGMGASHVRSLVAEGAKVVFGDILDDEGKAV
Mflv_4505|M.gilvum_PYR-GCK MTGR---LAGKVALISGAARGMGASHARVMAGHGAKVVCGDILDSDGEAV
MSMEG_3515|M.smegmatis_MC2_155 -MGR---VAGKVALISGGARGMGAAHARALVAEGAKVVIGDILDDEGAAL
Mvan_3000|M.vanbaalenii_PYR-1 -MGR---VDGKVALISGGAQGMGAADARALIAEGAKVVIGDILDDKGKAL
MUL_1037|M.ulcerans_Agy99 -MGR---VDGKVALISGGARGMGASHARLLVQEGAKVVIGDILDEEGKAL
MAB_4680c|M.abscessus_ATCC_199 MNGNSALLQGKNVIVTGGARGLGAAFSRHIVSQGGRVVIADVLDGDGRQL
TH_4254|M.thermoresistible__bu MTAGSTRLAGKVVLVTGAGGGLGTAIARRLADEGARLVLADLQESQCAAL
MLBr_01094|M.leprae_Br4923 -MEG---FAGKVAVVTGAGSGIGQALAIELARSGAKLAISDVDGEGLAQT
. ** .:::*.. *:* : : *.::. .*:
Mb2025|M.bovis_AF2122/97 AAELA----DAARYVHLDVTQPAQWTAAVDTAVTAFGGLHVLVNNAGILN
Rv2002|M.tuberculosis_H37Rv AAELA----DAARYVHLDVTQPAQWTAAVDTAVTAFGGLHVLVNNAGILN
MMAR_2981|M.marinum_M AAELG----DAARYVHLDVTQPEQWTAAVDTAVNEFGGLHVLVNNAGILN
MAV_2509|M.avium_104 AAEVG----EATRYLHLDVTKPEDWDAAVATALAEFGRIDVLVNNAGIIN
Mflv_4505|M.gilvum_PYR-GCK AAELG----DAARYVHLDVTSPDDWDRAVAAAVADFGGLDVLVNNAGILN
MSMEG_3515|M.smegmatis_MC2_155 AAELG----DAARYVHLDVTQAEDWDTAVATATTDFGLLNVLINNAGIVA
Mvan_3000|M.vanbaalenii_PYR-1 ADEINAETPDSIRYVHLDVTQADQWEAAVATAVDAFGKLNVLVNNAGTVA
MUL_1037|M.ulcerans_Agy99 AEEIG----DAARYVHLDVTQPDQWEAAVATAVDEFGKLDVLVNNVGIVA
MAB_4680c|M.abscessus_ATCC_199 AAELG----DAARFTHLDVTDAEQWRELIDTTLAGFGTITGLVNNAGISS
TH_4254|M.thermoresistible__bu ADELPAP-AGAHHPLGFDVADETAWISAISTVSAQYGALHALVNNAAIGA
MLBr_01094|M.leprae_Br4923 EGQLKAIG-ASARTDRLDVTEREAFLTYADVVHENFGKVNQIYNNAGIAF
:: : : :**:. : .. :* : : **..
Mb2025|M.bovis_AF2122/97 IGTIEDYALTEWQRILDVNLTGVFLGIRAVVKPMKEAGRGSIINISSIEG
Rv2002|M.tuberculosis_H37Rv IGTIEDYALTEWQRILDVNLTGVFLGIRAVVKPMKEAGRGSIINISSIEG
MMAR_2981|M.marinum_M IGTIEDYALSEWQRILDINVTGVFLGIRAAVKPMKEAGRGSIINISSIEG
MAV_2509|M.avium_104 IGTLEDYALSEWQRILDINLTGVFLGIRAVVKPMKKAGRGSIINISSIEG
Mflv_4505|M.gilvum_PYR-GCK IGTVEDYELSEWHRILDVNLTGVFLGIRAATPTMKAAGRGSIINIASIEG
MSMEG_3515|M.smegmatis_MC2_155 LGAIGKFDMTKWQNVIDVNLTGTFLGMQASVGAMKAAGGGSIINVSSIEG
Mvan_3000|M.vanbaalenii_PYR-1 LGQIGQFDMAKWQKVIDVNLTGTFLGMQASVEAMKTAGGGSIINISSIEG
MUL_1037|M.ulcerans_Agy99 LGQLKKFDLGKWQKVIDVNLTGTFLGMRAAVEPMTAAGSGSIINVSSIEG
MAB_4680c|M.abscessus_ATCC_199 AAMVADEPLEHFRKVIEINLVGVFIGMQAVIPAMRAHGCGSIVNISSAAG
TH_4254|M.thermoresistible__bu LGNVAEERLERWDRILDVGATGVWLGMKHAVPLIRRSGGGSVVNICSIFG
MLBr_01094|M.leprae_Br4923 TGDVEVSHFKDIERVMDVDYWGVVNGTKAFLPYLISSGDGHVINISSVFG
. : : .::::. *. * : : * * ::*:.* *
Mb2025|M.bovis_AF2122/97 LAG-TVACHGYTATKFAVRGLTKSTALELGPSG--IRVNSIHPGLVKTPM
Rv2002|M.tuberculosis_H37Rv LAG-TVACHGYTATKFAVRGLTKSTALELGPSG--IRVNSIHPGLVKTPM
MMAR_2981|M.marinum_M LAG-TIASHGYTVSKFAVRGLTKSTALELGPSG--IRVNSIHPGLVKTPM
MAV_2509|M.avium_104 MAG-TIACHGYTATKFAVRGLTKSAALELGPSG--IRVNSIHPGLIKTPM
Mflv_4505|M.gilvum_PYR-GCK MAG-TVGCHGYTATKFAVRGLTKSTALELGPFG--IRVNSVHPGLVKTPM
MSMEG_3515|M.smegmatis_MC2_155 LRG-APMVHPYVASKWAVRGLTKSAALELGAHQ--IRVNSIHPGFIRTPM
Mvan_3000|M.vanbaalenii_PYR-1 LRG-AVMVHPYVASKWAVRGLTKSAALELGSHN--IRVNSVHPGFIRTPM
MUL_1037|M.ulcerans_Agy99 LRG-APAVHPYVASKWAVRGLTKSAALELAPLN--IRVNSIHPGFIRTPM
MAB_4680c|M.abscessus_ATCC_199 LVG-LALTSGYGASKWGVRGLSKVAAIELGRER--IRVNSVHPGVIYTPM
TH_4254|M.thermoresistible__bu TVGGFGDSAAYHAAKGAVRTLTKNAALHWAPDG--VRVNSLHPGFIETPQ
MLBr_01094|M.leprae_Br4923 LFS-VPGQAAYNSAKFAVRGFTEALREEMALAGRPVNVTTVYPGGIKTAI
. * :* .** ::: . . :.*.:::** : *.
Mb2025|M.bovis_AF2122/97 TDWV----------PEDIFQTALGRAAEPVEVSNLVVYLASDESSYSTGA
Rv2002|M.tuberculosis_H37Rv TDWV----------PEDIFQTALGRAAEPVEVSNLVVYLASDESSYSTGA
MMAR_2981|M.marinum_M TEWV----------PEDLFQTALGRAAEPMEVSNLVVYLASDESSYSTGA
MAV_2509|M.avium_104 TEWV----------PEDIFQSALGRAAEPKEVSNLVVYLASDESSYSTGS
Mflv_4505|M.gilvum_PYR-GCK ADWV----------PEDIFQSALGRIAQPHEVSNLVVYLASDESSYSTGA
MSMEG_3515|M.smegmatis_MC2_155 TEHF----------PDDMLRIPLGRPGEVDEVSSFVVFLASDESRYATGA
Mvan_3000|M.vanbaalenii_PYR-1 TKHF----------PDNMLRIPLGRPGQPEEVATFVVFLASDESRYATGA
MUL_1037|M.ulcerans_Agy99 TANL----------PDDMVTIPLGRPAESREVSTFVVFLASDDASYATGS
MAB_4680c|M.abscessus_ATCC_199 TAKEGF-----REGEGNMPIAAMGRVGNADEVAGAVAYLLSDAASYVTGA
TH_4254|M.thermoresistible__bu LRELYKGT---ERYERMLAGTPLGRLGRPDEIAAVVAFLVSDDASFITGS
MLBr_01094|M.leprae_Br4923 ARNATAAEGLDVSKIASRFDTWVAHTSPQHAARIILKAVRKKKARVLVGP
:.: . : : .. : .*.
Mb2025|M.bovis_AF2122/97 EFVVDGGTVAGLAHNDFGAVEVSSQPEWVT---
Rv2002|M.tuberculosis_H37Rv EFVVDGGTVAGLAHNDFGAVEVSSQPEWVT---
MMAR_2981|M.marinum_M EFVVDGGTVAGLAHKDFSAVDVAAQPEWVT---
MAV_2509|M.avium_104 EFVVDGGTTAGLGHKDFSNVETDAQPDWVT---
Mflv_4505|M.gilvum_PYR-GCK EFVVDGGTLAGLAHKDFSAVDVDQQPEWIT---
MSMEG_3515|M.smegmatis_MC2_155 EFVMDGGLVNDVPHKL-----------------
Mvan_3000|M.vanbaalenii_PYR-1 EFVMDGGLTNDVPHK------------------
MUL_1037|M.ulcerans_Agy99 EFVMDGGLVTDVPHKQF----------------
MAB_4680c|M.abscessus_ATCC_199 ELAVDGGWTAGPTLTTITGQ-------------
TH_4254|M.thermoresistible__bu EVYADGGWTAR----------------------
MLBr_01094|M.leprae_Br4923 DAKVANVVVRFSGGAGYQRLFAQVASRLILNQR
: .