For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AVSRFSLEGRVAVITGGGTGIGRASALVLAEHGADIVLAGRRLEPLKNTATEIEALGRRALAVPTDVTDP EACRSLVDATVDEFGRLDILLNNAGGGETKSLMKWTDDEWRQVLDLNLSSAWYLSRAAVKPMLSQGKGSI VNISSGASLLAMPQAAIYGAAKAGLNNLTGSMAAAWTRKGVRVNCIACGAIRTPGLEADAARQGFDIDMI GQTNGSGRIAEPDEIGYGVLFFASDASSYCSGQTLYMHGGPGPAGV*
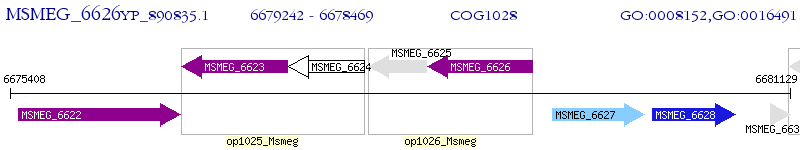
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6626 | - | - | 100% (257) | short-chain dehydrogenase/reductase SDR |
| M. smegmatis MC2 155 | MSMEG_4074 | - | 6e-46 | 41.46% (246) | peroxisomal trans-2-enoyl-CoA reductase |
| M. smegmatis MC2 155 | MSMEG_4117 | - | 2e-41 | 37.14% (245) | 2-deoxy-D-gluconate 3-dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0950c | - | 1e-37 | 40.08% (247) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1263 | - | 1e-132 | 89.72% (253) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv0927c | - | 1e-37 | 40.08% (247) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01740 | - | 2e-22 | 32.45% (188) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0833 | - | 1e-37 | 37.10% (248) | Short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_4581 | - | 2e-39 | 40.08% (252) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_1391 | - | 1e-115 | 77.87% (253) | short-chain dehydrogenase/reductase SDR |
| M. thermoresistible (build 8) | TH_4266 | - | 1e-129 | 88.93% (253) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_4442 | - | 2e-38 | 39.29% (252) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4921 | - | 4e-37 | 38.89% (252) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1263|M.gilvum_PYR-GCK -----------MSDRFSMEGRVAVITGGGTGIGRASALVLAEHGADVVLA
TH_4266|M.thermoresistible__bu -----------VDDRFSMAGRVAVITGGGTGIGRASALVLAEYGADIVLA
MSMEG_6626|M.smegmatis_MC2_155 ----------MAVSRFSLEGRVAVITGGGTGIGRASALVLAEHGADIVLA
MAV_1391|M.avium_104 -----------MSDRFALTDRVAVITGAGTGIGRASALVLAEHGADIVLA
Mb0950c|M.bovis_AF2122/97 ----------MILDMFRLDDKVAVITGGGRGLGAAIALAFAQAGADVLIA
Rv0927c|M.tuberculosis_H37Rv ----------MILDMFRLDDKVAVITGGGRGLGAAIALAFAQAGADVLIA
MMAR_4581|M.marinum_M ----------MILDRFRLDDQVAVITGGGRGLGAAIALAFAEAGADVVIA
MUL_4442|M.ulcerans_Agy99 ----------MILDRFRLDDQVAVITGGGRGFGAAIALAFAEAGADVVIA
Mvan_4921|M.vanbaalenii_PYR-1 MTRGRQREANVILDRFRLDDQVAVVTGAGRGLGAAMALAFAEAGADVLIA
MAB_0833|M.abscessus_ATCC_1997 ------MNRETFSRLFDLTGRTAIVTGGTRGIGRALAEGYACAGANVVVA
MLBr_01740|M.leprae_Br4923 ----------MVKTAQYFVGKRCFVTGAASGIGRATALRLAAYGAELYLT
: .: ..:**. *:* * * * **:: ::
Mflv_1263|M.gilvum_PYR-GCK GRRAEPLKETAAEVEALGRRA-LAVPTDVTDADACQALVDTTLAEFGRLD
TH_4266|M.thermoresistible__bu GRRPEPLKKTAAEIEALGRRA-LAVPTDVTAPDECRALVDATVGEFGRLD
MSMEG_6626|M.smegmatis_MC2_155 GRRLEPLKNTATEIEALGRRA-LAVPTDVTDPEACRSLVDATVDEFGRLD
MAV_1391|M.avium_104 GRRPDPLQATAKEVEALGRRS-LIVPTDVTETQQCQHLVDATMAEFGRLD
Mb0950c|M.bovis_AF2122/97 SRTSSELDAVAEQIRAAGRRA-HTVAADLAHPEVTAQLAGQAVGAFGKLD
Rv0927c|M.tuberculosis_H37Rv SRTSSELDAVAEQIRAAGRRA-HTVAADLAHPEVTAQLAGQAVGAFGKLD
MMAR_4581|M.marinum_M SRTQSQLDEVADTVRSIGRRA-HVVAADLAHPEATAELAGQAVEAFGRLD
MUL_4442|M.ulcerans_Agy99 SRTQSQLDEVADTVRSIGRRA-HVVAADLAHPEATAELAGQAVEAFGRLD
Mvan_4921|M.vanbaalenii_PYR-1 ARTRSQLEEVAEQIEGTGRRA-HIVVADLAHAEDTAALAAGAVEAFGRLD
MAB_0833|M.abscessus_ATCC_1997 SRKPEACTEAVERLRSLGARA-LGVPTHTGDIHSLEELVRATAEEFGGID
MLBr_01740|M.leprae_Br4923 DRHCDGLAQTVSEARALGAQVPEYRALDISDYDEVAAFGADIHARHPSMD
* . .. .. * : . . : . :*
Mflv_1263|M.gilvum_PYR-GCK VLLNNAGGGETKSLMKWTDDEWHHVLDLNLSSAWYLSRAAAKPMIAQG-S
TH_4266|M.thermoresistible__bu VLVNNAGGGATKPLMKWTDDEWHEVLNLNLSSAWYLSRAAAKPMIDQG-R
MSMEG_6626|M.smegmatis_MC2_155 ILLNNAGGGETKSLMKWTDDEWRQVLDLNLSSAWYLSRAAVKPMLSQG-K
MAV_1391|M.avium_104 ILVNNAGGGETKGITRWTEEEWHDVVDLNLGSVWFLSRCAVKPMMTQG-S
Mb0950c|M.bovis_AF2122/97 IVVNNVGGTMPNTLLSTSTKDLADAFAFNVGTAHALTVAAVPLMLEHSGG
Rv0927c|M.tuberculosis_H37Rv IVVNNVGGTMPNTLLSTSTKDLADAFAFNVGTAHALTVAAVPLMLEHSGG
MMAR_4581|M.marinum_M IVVNNVGGTMPNTLLTTSTKDLKDAFTFNVATAHALTVAAVPLMLEHSGG
MUL_4442|M.ulcerans_Agy99 IVVNNVGGTMPNTLLTTSTKDLKDAFTFNVATAHALTVAAVPLMLEHSGG
Mvan_4921|M.vanbaalenii_PYR-1 IVVNNVGGTMPNALLTTSTKDMRDAFAFNVATAHALTTAAVPLMLEHSGG
MAB_0833|M.abscessus_ATCC_1997 IVVNNAANALTEPVGAFTVDGWDKSFGVNLRGPVFLVQAALPHLVASP-H
MLBr_01740|M.leprae_Br4923 IVMNIAGVSVWGTVDRLTHDQWSNVVAINLMGPIHIIETFVPAILAAGRG
:::* .. : : . . . .*: : ::
Mflv_1263|M.gilvum_PYR-GCK GAIVNISSGAGLLAMPQAPIYGAAKAGLQNLTGSMAAAWTRKGVRVNCIA
TH_4266|M.thermoresistible__bu GAIVNISSGAGLLAMPQAPVYAAAKAGLNNLTGSMAAAWTRKGVRVNCIA
MSMEG_6626|M.smegmatis_MC2_155 GSIVNISSGASLLAMPQAAIYGAAKAGLNNLTGSMAAAWTRKGVRVNCIA
MAV_1391|M.avium_104 GAIVNISSGASLIAMPQAAIYAAAKAGVNNLTGSMAAAWGRKGIRVNCIA
Mb0950c|M.bovis_AF2122/97 GSVINISSTMGRLAARGFAAYGTAKAALAHYTRLAALDLCPR-VRVNAIA
Rv0927c|M.tuberculosis_H37Rv GSVINISSTMGRLAARGFAAYGTAKAALAHYTRLAALDLCPR-VRVNAIA
MMAR_4581|M.marinum_M GSIINITSTMGRVAGRGFAAYGTAKAALAHYTRLAALDLCPR-IRVNAIA
MUL_4442|M.ulcerans_Agy99 GGIINITSTMGRVAGRGFAAYGTAKVALAHYTRLAALDLCPR-IRVNAIA
Mvan_4921|M.vanbaalenii_PYR-1 GSIINITSTMGRLAGRGFAAYGTAKAALAHYTRLSALDLAPR-IRVNAIA
MAB_0833|M.abscessus_ATCC_1997 AAVLNVVSVGAFMFGQGVSMYSAAKAALVSYTRSAAAELASRGVRVNALA
MLBr_01740|M.leprae_Br4923 GHLVNVSSAAGLVALPWHAAYSASKYGLRGLSEVLRFDLARYRIGVSVVV
. ::*: * . : . *.::* .: : : *. :.
Mflv_1263|M.gilvum_PYR-GCK CGAVRTPGLEADAKRQG--FDIDMIGQTNGSGRIAEPDEIGYGVLFFASD
TH_4266|M.thermoresistible__bu CGAIRTPGLEADAKRQG--FDIDMIGQTNGSGRIAEPDEIGYGVLFFASD
MSMEG_6626|M.smegmatis_MC2_155 CGAIRTPGLEADAARQG--FDIDMIGQTNGSGRIAEPDEIGYGVLFFASD
MAV_1391|M.avium_104 CGAIRTDGLLADAAKGG--FDVDQLGAMNAMGRIAEPDEIGYGVLFFASD
Mb0950c|M.bovis_AF2122/97 PGSILTSALEVVAAND---ELRAPMEQATPLRRLGDPVDIAAAAVYLASP
Rv0927c|M.tuberculosis_H37Rv PGSILTSALEVVAAND---ELRAPMEQATPLRRLGDPVDIAAAAVYLASP
MMAR_4581|M.marinum_M PGSILTSALEVVAAND---ELRAPMEQATPLRRLGDPVDIAAAAVYLASP
MUL_4442|M.ulcerans_Agy99 PGSILTSALEVVAAND---ELRAPMEQATPLRRLGDPVDIAAAAVYLASP
Mvan_4921|M.vanbaalenii_PYR-1 PGSILTSALEIVADND---ELRGPMEKATPLRRLGDPLDIAAAAVYLASP
MAB_0833|M.abscessus_ATCC_1997 PGAIDTDMVRKTPPAE---VER--MANTNHLKRLGLPDEMVGTALLLTSD
MLBr_01740|M.leprae_Br4923 PGAVRTPLVDTIEIAGVDRQDPRFSRWVDRFRGHAVSAERAAEKILAGVA
*:: * : . . : :
Mflv_1263|M.gilvum_PYR-GCK ASSYCSGQTLYMHGGPGPAGV-----------------------------
TH_4266|M.thermoresistible__bu ASSYCSGQTLYMHGGPGPAGV-----------------------------
MSMEG_6626|M.smegmatis_MC2_155 ASSYCSGQTLYMHGGPGPAGV-----------------------------
MAV_1391|M.avium_104 ASSYCSGQTLYIHGGPGPAGI-----------------------------
Mb0950c|M.bovis_AF2122/97 AGSFLTGKTLEVDGGLTFPNLDLPIPDL----------------------
Rv0927c|M.tuberculosis_H37Rv AGSFLTGKTLEVDGGLTFPNLDLPIPDL----------------------
MMAR_4581|M.marinum_M AGSFLTGKTLEVDGGLTHPNLDIPVPDL----------------------
MUL_4442|M.ulcerans_Agy99 AGSFLTGKTLEVDGGLTHPNLDIPVPDL----------------------
Mvan_4921|M.vanbaalenii_PYR-1 AGSYLTGKTLEVDGGITFPNLDLPIPDL----------------------
MAB_0833|M.abscessus_ATCC_1997 AGSYITGQTIIADGGLVAR-------------------------------
MLBr_01740|M.leprae_Br4923 KNRYLIYTSHDIRVLYGFKRLAWWPYGVVMKQVNVVFTRALRPSPATMRC
. : :
Mflv_1263|M.gilvum_PYR-GCK ----------------------
TH_4266|M.thermoresistible__bu ----------------------
MSMEG_6626|M.smegmatis_MC2_155 ----------------------
MAV_1391|M.avium_104 ----------------------
Mb0950c|M.bovis_AF2122/97 ----------------------
Rv0927c|M.tuberculosis_H37Rv ----------------------
MMAR_4581|M.marinum_M ----------------------
MUL_4442|M.ulcerans_Agy99 ----------------------
Mvan_4921|M.vanbaalenii_PYR-1 ----------------------
MAB_0833|M.abscessus_ATCC_1997 ----------------------
MLBr_01740|M.leprae_Br4923 VEPIKVGVYSQPRSGEGGKSGN