For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLAAALMFPVVGGFGLMSNRASDVVANGSAALVEGEVPQVSTMVDAKGNTIAWLYSQRRFEVPSEQIANT MKLAIVSIEDKRFAEHNGVDWQGTLTGLSGYLSGNPDTRGGSTIEQQYVKNYQLLVVAQTDAERRAAIET TPARKLREIRMALTLDKTFTKPEILTRYLNLVSFGNGAYGIQDAAQTYFGVNASELNWQQAALLAGMVQS TSALNPYTNPDGALARRNLVLDTMIQNIPQEAEALRAAKEQPLGILPQPNELPRGCIAAGDRAFFCDYVL EYLARAGISKEQVAKGGYLIKTTLDPDVQSSVKSAIDSIAAPDTPGVASVMSVIKPGKESHPVLAMASNR TYGLDTERGETMQPQPFSLVGNGAGSVFKIFTVAAAMEMGMGINTQLPVPARFEAKGLGSGGARGCPPAT WCVQNAGGYRGSMSVTDALATSPNTAFAKLISQIGVQRAVDMAVRLGLRSYAQPGTARDYDPESSESLAD FVKRQNIGSFTLGPIEVNALELSNVAATLASGGTWCPPNPIAQVVDRNGNEVAVTTETCEQVVPEGLANT LANAMSKDDTGSGTAAASAGSVGWNLPMSGKTGTTEDNRSSAFLGFTNQYAAANYIYDDSPNPSELCSFP LRQCGSGNLFGGNEPARTWFTAMKPIAESFGPVALPPTDPRYVDGGPGSRVPSVSGLTEEAARQRLKEAG FQVADQASYVNSGARSGTVVGTTPSGQTIPGSIITIQLSNGIPPPPPPPVNIPLPGGPPEIGQTVVEIPG LPPITVPVLGPPPPPPPP
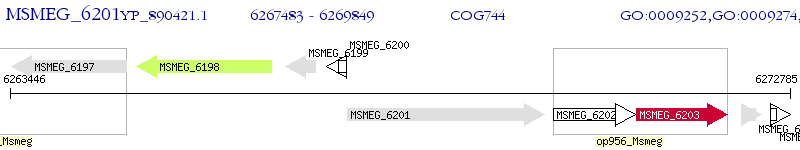
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6201 | - | - | 100% (788) | transglycosylase |
| M. smegmatis MC2 155 | MSMEG_4384 | - | 0.0 | 72.19% (791) | penicillin binding protein transpeptidase domain-containing |
| M. smegmatis MC2 155 | MSMEG_6900 | - | 4e-50 | 26.30% (768) | penicillin-binding protein 1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3707 | ponA2 | 0.0 | 80.91% (791) | bifunctional membrane-associated penicillin-binding protein |
| M. gilvum PYR-GCK | Mflv_1365 | - | 0.0 | 85.19% (790) | glycosyl transferase family protein |
| M. tuberculosis H37Rv | Rv3682 | ponA2 | 0.0 | 80.91% (791) | bifunctional membrane-associated penicillin-binding protein |
| M. leprae Br4923 | MLBr_02308 | pon1 | 0.0 | 79.16% (782) | penicillin binding protein (class A) |
| M. abscessus ATCC 19977 | MAB_0408c | - | 0.0 | 74.39% (785) | bifunctional membrane-associated penicillin-binding protein |
| M. marinum M | MMAR_5171 | ponA2 | 0.0 | 81.89% (784) | bifunctional membrane-associated penicillin- binding |
| M. avium 104 | MAV_0446 | - | 0.0 | 81.24% (789) | transglycosylase |
| M. thermoresistible (build 8) | TH_2122 | ponA | 0.0 | 87.12% (730) | PROBABLE BIFUNCTIONAL MEMBRANE-ASSOCIATED PENICILLIN-BINDING |
| M. ulcerans Agy99 | MUL_4257 | ponA2 | 0.0 | 81.63% (784) | bifunctional membrane-associated penicillin- binding |
| M. vanbaalenii PYR-1 | Mvan_5442 | - | 0.0 | 85.68% (789) | glycosyl transferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6201|M.smegmatis_MC2_155 -----------------------------MLAAALMFPVVGGFGLMSNRA
TH_2122|M.thermoresistible__bu MPEDPPTAPTESTRAATVIKLAWCCLLAAVIAAALMFPVVGGIGLLSNRA
Mflv_1365|M.gilvum_PYR-GCK MPEQPPTQP---PRAVTVIKLAWCVVLASVIVAGLLFPVVGGFGLVSNRA
Mvan_5442|M.vanbaalenii_PYR-1 MPEHPSGPP---PRAVTVIKLAWCVLLASVVAAGLMFPVVGGFGLMPNRA
Mb3707|M.bovis_AF2122/97 MPERL-------PAAITVLKLAGCCLLASVVATALTFPFAGGLGLMSNRA
Rv3682|M.tuberculosis_H37Rv MPERL-------PAAITVLKLAGCCLLASVVATALTFPFAGGLGLMSNRA
MMAR_5171|M.marinum_M MSERP-------PAALTLLKLSGFCLLASIVAAALMFPFAGGIGLMSNRA
MUL_4257|M.ulcerans_Agy99 MSERP-------PAALSLLKLSGFCLLASIVAAALMFPFAGGIGLMSNRA
MAV_0446|M.avium_104 ------------------------------MATALLFPVAGGIGLVSNRA
MLBr_02308|M.leprae_Br4923 MSERL-------PAVLAVLKLAGYCLLAGVVTTAMMFPLAGGLGVISNRA
MAB_0408c|M.abscessus_ATCC_199 MPERLSP---QIPPSVTLAKLAGCGLVAAVILAGLMFPFAGGFGLASNRA
: :.: **..**:*: .***
MSMEG_6201|M.smegmatis_MC2_155 SDVVANGSAALVEGEVPQVSTMVDAKGNTIAWLYSQRRFEVPSEQIANTM
TH_2122|M.thermoresistible__bu SDVVANGSAQLVEGDVPAVSTMVDAEGNPIAWLYSQRRWEVPSEEIADTM
Mflv_1365|M.gilvum_PYR-GCK SDVVANGSAQLVEGEVPQVSTMTDSKGNIIAWLYSQRRFEVPSDQIANTM
Mvan_5442|M.vanbaalenii_PYR-1 SDVVANGSAQLVEGEVPQVSTMTDAKGNVIAWLYNQRRFEVPSDQIANTM
Mb3707|M.bovis_AF2122/97 SEVVANGSAQLLEGQVPAVSTMVDAKGNTIAWLYSQRRFEVPSDKIANTM
Rv3682|M.tuberculosis_H37Rv SEVVANGSAQLLEGQVPAVSTMVDAKGNTIAWLYSQRRFEVPSDKIANTM
MMAR_5171|M.marinum_M SEVVANGSAQLLQGEVPAVSTMVDAKGNTIAWLYSQRRFEVPSDKIANTM
MUL_4257|M.ulcerans_Agy99 SEVVANGSAQLLQGEVPAVSTMVDAKGNTIAWLYSQRRFEVPSDKIANTM
MAV_0446|M.avium_104 SEVVANGSAQLLEGEVPAVSTMVDAKGNTIAWLYSQRRFEVPSDKIANTM
MLBr_02308|M.leprae_Br4923 SEVVANGSAQLLEGEVPAVSTMVDAKGNTIAWLYSQRRFEVPTDKIANTM
MAB_0408c|M.abscessus_ATCC_199 SDVVANGSTQLLQGEVPAVTTVEDAKGNKIAYLYTQRRFEVSADEIADTM
*:******: *::*:** *:*: *::** **:**.***:**.:::**:**
MSMEG_6201|M.smegmatis_MC2_155 KLAIVSIEDKRFAEHNGVDWQGTLTGLSGYLSGNPDTRGGSTIEQQYVKN
TH_2122|M.thermoresistible__bu KLAIVAIEDKRFAEHNGVDWQGTLTGLTGYLRGDSDTRGGSTLEQQYVKN
Mflv_1365|M.gilvum_PYR-GCK KLAIVSIEDKRFADHNGVDWQGTLTGLSGYLSGNLDTRGGSTIEQQYVKN
Mvan_5442|M.vanbaalenii_PYR-1 KLAIVSIEDKRFADHNGVDWQGTLTGLSGYLSGNLDTRGGSTIEQQYVKN
Mb3707|M.bovis_AF2122/97 KLAIVSIEDKRFADHSGVDWKGTLTGLAGYASGDLDTRGGSTLEQQYVKN
Rv3682|M.tuberculosis_H37Rv KLAIVSIEDKRFADHSGVDWKGTLTGLAGYASGDLDTRGGSTLEQQYVKN
MMAR_5171|M.marinum_M KLAIVSIEDKRFAEHNGVDWKGTLTGLAGYASGDLDTRGGSTIEQQYIKN
MUL_4257|M.ulcerans_Agy99 KLAIVSIEDKRFAEHNGVDWKGTLTGLAGYASGDLDTRGGSTIEQQYIKN
MAV_0446|M.avium_104 KLAIVSIEDKRFADHNGVDWKGTLTGLAGYARGDVDTRGGSTIEQQYIKN
MLBr_02308|M.leprae_Br4923 KLAIVSIEDKRFTDHNGVDWPGTLTGLAGYASGDVDTRGGSTIEQQYVKN
MAB_0408c|M.abscessus_ATCC_199 KLALLSIEDKRFTSHNGVDWKGTLGGLAGYASGDEDTRGGSTIEQQYVKN
***:::******:.*.**** *** **:** *: *******:****:**
MSMEG_6201|M.smegmatis_MC2_155 YQLLVVAQTDAERRAAIETTPARKLREIRMALTLDKTFTKPEILTRYLNL
TH_2122|M.thermoresistible__bu YQLLVVAQTDAEKRAAIETTPARKLREIRMALTLDKTLTKPEILTRYLNL
Mflv_1365|M.gilvum_PYR-GCK YQLLVIAQTDAEKRAAIETTPARKLREIRMALTLDKTFTKPEILTRYLNL
Mvan_5442|M.vanbaalenii_PYR-1 YQLLVIAQTDAEKRAAIETTPARKLREIRMALTLDKTFTKPEILTRYLNL
Mb3707|M.bovis_AF2122/97 YQLLVTAQTDAEKRAAVETTPARKLREIRMALTLDKTFTKSEILTRYLNL
Rv3682|M.tuberculosis_H37Rv YQLLVTAQTDAEKRAAVETTPARKLREIRMALTLDKTFTKSEILTRYLNL
MMAR_5171|M.marinum_M YQLLVTAQTDAEKRAAVETTPARKLREIRMALTLDKTFTKPEILTRYLNL
MUL_4257|M.ulcerans_Agy99 YQLLVTAQTDAEKRAAVETTPARKLREIRMALTLDKTFTKPEILTRYLNL
MAV_0446|M.avium_104 YQLLVTAKTDAEKRAAVETTPARKLREIRMALTLDKTFTKPEILTRYLNL
MLBr_02308|M.leprae_Br4923 YQLLVTAQTDAEKRAAVETTPARKLREIRIALTLDKTFTKPEILTRYLNL
MAB_0408c|M.abscessus_ATCC_199 YQLLVVAKTDAERKAAIETTPARKLREIRMALRLDQTFTKQEILTRYLNL
***** *:****::**:************:** **:*:** *********
MSMEG_6201|M.smegmatis_MC2_155 VSFGNGAYGIQDAAQTYFGVNASELNWQQAALLAGMVQSTSALNPYTNPD
TH_2122|M.thermoresistible__bu VSFGNGAYGIQDAAQTYFGINASELDWHQAALLAGLVQSTTALNPYTNPD
Mflv_1365|M.gilvum_PYR-GCK VSFGNNAFGVQDAAQTYFGINATDLNWQQAALLAGMVQSTSTLNPYTNPD
Mvan_5442|M.vanbaalenii_PYR-1 VSFGNNAFGVQDAAQTYFGVNASELNWQQAALLAGMVQSTSTLNPYTNPD
Mb3707|M.bovis_AF2122/97 VSFGNNSFGVQDAAQTYFGINASDLNWQQAALLAGMVQSTSTLNPYTNPD
Rv3682|M.tuberculosis_H37Rv VSFGNNSFGVQDAAQTYFGINASDLNWQQAALLAGMVQSTSTLNPYTNPD
MMAR_5171|M.marinum_M VSFGNNSFGVQDAAQTYFGINASELNWQQSALLAGMVQSTSTLNPYTNPD
MUL_4257|M.ulcerans_Agy99 VSFGNNSFGVQDAAQTYFGINASELNWQQSALLAGMVQSTSTLNPYTNPD
MAV_0446|M.avium_104 VSFGNNSFGVQDAAQTYFGINASDLNWQQAALLAGMVQSTSALNPYTNPE
MLBr_02308|M.leprae_Br4923 VSFGNNSFGVQDAAQTYFGVNASDLNWQQAALLAGMVQSTSTLNPYTNPE
MAB_0408c|M.abscessus_ATCC_199 VSFGNNSFGVQDAAQTYFGVDAKDLNWQQAALLAGMVQSTSGLNPYVNPE
*****.::*:*********::*.:*:*:*:*****:****: ****.**:
MSMEG_6201|M.smegmatis_MC2_155 GALARRNLVLDTMIQNIPQEAEALRAAKEQPLGILPQPNELPRGCIAAGD
TH_2122|M.thermoresistible__bu GALARRNLVLDTMIDQLPQEAEALRAAKEQPLGILPRPNELPRGCIAAGD
Mflv_1365|M.gilvum_PYR-GCK GALARRNVVLDTMIENLPEYADELRAARQEPLGVLPQPKELPRGCIAAGD
Mvan_5442|M.vanbaalenii_PYR-1 GALARRNVVLDTMIENLPEHADELRTAKQQPLGILPQPKELPRGCIAAGD
Mb3707|M.bovis_AF2122/97 GALARRNVVLDTMIENLPGEAEALRAAKAEPLGVLPQPNELPRGCIAAGD
Rv3682|M.tuberculosis_H37Rv GALARRNVVLDTMIENLPGEAEALRAAKAEPLGVLPQPNELPRGCIAAGD
MMAR_5171|M.marinum_M GALARRNLVLDTMIQNIPSEADALRAAKAEPLGILPQPNELPRGCIAAGD
MUL_4257|M.ulcerans_Agy99 GALARRNLVLDTMIQNIPSEADALRAAKAEPLGILPQPNELPRGCIAAGD
MAV_0446|M.avium_104 GALARRNLVLDTMIENIPQEADALRAAKAAPLGILPQPNELPRGCIAAGD
MLBr_02308|M.leprae_Br4923 GALARRNLVLDTMIENLPQDAEALRAAKTEPLGILPRPNELPRGCIAAGD
MAB_0408c|M.abscessus_ATCC_199 RALARRNLVLDTMIANSPDKKGELLAAKAQPLGVLPQPNELPRGCIAAGD
******:****** : * * :*: ***:**:*:***********
MSMEG_6201|M.smegmatis_MC2_155 RAFFCDYVLEYLARAGISKEQVAKGGYLIKTTLDPDVQSSVKSAIDSIAA
TH_2122|M.thermoresistible__bu RAFFCDYVLEYLARAGISKEQVAKGGYLIKTTLDPEVQRAVKSAIDQYAR
Mflv_1365|M.gilvum_PYR-GCK RAFFCDYALDYLARAGISKDQVAKGGYMIKTTLDPDVQASVKNAITQYAS
Mvan_5442|M.vanbaalenii_PYR-1 RAFFCDYALDYLARAGISKDQVAKGGYLIKTTLEPDVQASVKNAINEFAS
Mb3707|M.bovis_AF2122/97 RAFFCDYVQEYLSRAGISKEQVATGGYLIRTTLDPEVQAPVKAAIDKYAS
Rv3682|M.tuberculosis_H37Rv RAFFCDYVQEYLSRAGISKEQVATGGYLIRTTLDPEVQAPVKAAIDKYAS
MMAR_5171|M.marinum_M RAFFCDYVQEYLSRAGISKEQLARGGYLIRTTLDPDVQVPVKAAIDKFAS
MUL_4257|M.ulcerans_Agy99 RAFFCDYVQEYLSRAGISKEQLARGGYLIRTTLDPDVQVPVKAAIDKFAS
MAV_0446|M.avium_104 RAYFCDYVQEYLSRAGISKEQVARGGYLIKTTLDPDVQIPVKAAIDKFAS
MLBr_02308|M.leprae_Br4923 RAFFCDYVQEYLSHAGISKDRLAKGGYLIRTTLDPNVQAPVKAAIDKFAS
MAB_0408c|M.abscessus_ATCC_199 RAFFCDYVLDYLAKSGISKDQVMKGGYLIRTTLDPDVQNSVKQAVSHVAD
**:****. :**:::****::: ***:*:***:*:** .** *: *
MSMEG_6201|M.smegmatis_MC2_155 PDTPGVASVMSVIKPGKESHPVLAMASNRTYGLDTERGETMQPQPFSLVG
TH_2122|M.thermoresistible__bu PDANGVASVMSVIRPGKESHPVLAMASSRHYGLNSEAGETMQPQPFSLVG
Mflv_1365|M.gilvum_PYR-GCK PDLPGVASVMSVVKPGKDSHPVLAMASNRTYGLNLDAGETMQPQPFSLVG
Mvan_5442|M.vanbaalenii_PYR-1 PDLPGVASVMSVIRPGKDSHPVLAMASNRTYGLNLDAGETMQPQPFSLVG
Mb3707|M.bovis_AF2122/97 PNLAGISSVMSVIKPGKDAHKVLAMASNRKYGLDLEAGETMRPQPFSLVG
Rv3682|M.tuberculosis_H37Rv PNLAGISSVMSVIKPGKDAHKVLAMASNRKYGLDLEAGETMRPQPFSLVG
MMAR_5171|M.marinum_M PTLPGISSVMSVIKPGKESHQVLAMASNRKYGLDINAGETMRPQPFSLVG
MUL_4257|M.ulcerans_Agy99 PTLPGISSVMSVIKPGKESHQVLAMASNRAYGLDINAGETMRPQPFSLVG
MAV_0446|M.avium_104 PTLPGISSVMSVIQPGKTSHKVMAMASNRTYGLDVDAGQTMRPQPFSLVG
MLBr_02308|M.leprae_Br4923 PNLAGISSVMSVITPGKDAHRVIAMGSNRRYGLDTEAGETMRPQTFSLVG
MAB_0408c|M.abscessus_ATCC_199 PGLNGIANVMSVIKPGQTSHPILAMASSRTYGLDGNAFQTMQPQPFSLVG
* *::.****: **: :* ::**.*.* ***: : :**:**.*****
MSMEG_6201|M.smegmatis_MC2_155 NGAGSVFKIFTVAAAMEMGMGINTQLPVPARFEAKGLGSG-GARGCPPAT
TH_2122|M.thermoresistible__bu HGAGSVFKIFTTAAAMEMGMGINTQLEAPAFFQAKGLGSS-DTPGCPAAT
Mflv_1365|M.gilvum_PYR-GCK NGAGSVFKIFTTAAAMEMGMGINAQLDSPSRFEAKGLGSG-GARGCPPAT
Mvan_5442|M.vanbaalenii_PYR-1 NGAGSVFKIFTTAAAMEMGMGINAQLDSPSRFEAKGLGSG-GARGCPPAT
Mb3707|M.bovis_AF2122/97 DGAGSIFKIFTTAAALDMGMGINAQLDVPPRFQAKGLGSG-GAKGCPKET
Rv3682|M.tuberculosis_H37Rv DGAGSIFKIFTTAAALDMGMGINAQLDVPPRFQAKGLGSG-GAKGCPKET
MMAR_5171|M.marinum_M DGAGSIFKIFTTAAALDMGMGINAQLDVPGRFQGKGLGSG-GAKGCPKDT
MUL_4257|M.ulcerans_Agy99 DGAGSIFKIFTTAAALDMGMGISAQLDVPGRFQGKGLGSG-GAKGCPKDT
MAV_0446|M.avium_104 DGAGSVFKIFTTAAALDMGMGINATLEVPGRFQAKGMGSG-GAKGCPKDT
MLBr_02308|M.leprae_Br4923 DGAGSVFKIFTTAAALDMGMGINAKLEVPSRFQAKGMGSG-GAKGCPKET
MAB_0408c|M.abscessus_ATCC_199 DGAGSIFKIFTVAAAMDMGMGINTVLDVPPTFAAKGLGSSDGSKGCPKDN
.****:*****.***::*****.: * * * .**:**. .: *** .
MSMEG_6201|M.smegmatis_MC2_155 WCVQNAGGYRGSMSVTDALATSPNTAFAKLISQIGVQRAVDMAVRLGLRS
TH_2122|M.thermoresistible__bu WCVRNAGSYRGSMNVTDALATSPNTAFAKLISQIGVPRTVDMAVRLGLRS
Mflv_1365|M.gilvum_PYR-GCK WCVQNAGNYRGTMSVTDALATSPNTAFAKLIAQVGVQRTVDMAVRLGLRS
Mvan_5442|M.vanbaalenii_PYR-1 WCVQNAGNYRGTMNVTDALATSPNTAFAKLIAQVGVQRTVDMAVKLGLRS
Mb3707|M.bovis_AF2122/97 WCVVNAGNYRGSMNVTDALATSPNTAFAKLISQVGVGRAVDMAIKLGLRS
Rv3682|M.tuberculosis_H37Rv WCVVNAGNYRGSMNVTDALATSPNTAFAKLISQVGVGRAVDMAIKLGLRS
MMAR_5171|M.marinum_M WCVINAGNYRGSMSVTDALATSPNTAFAKLISQVGVSRTVDMAIKLGLRS
MUL_4257|M.ulcerans_Agy99 WCVINAGNYRGSMSVTDALATSPNTAFAKLISQVGVSRTVDMAIKLGLRS
MAV_0446|M.avium_104 WCVVNAGNYKGSMNVTEALATSPNTAFAKLIQQVGVTRTVDMAIKLGLRS
MLBr_02308|M.leprae_Br4923 WCVINAGNYRGSMNVTDALATSPNTAFAKLIQQVGVARTVDMAIKLGLRS
MAB_0408c|M.abscessus_ATCC_199 WCVKNVGNYRSPMSVTDALATSPNTAFAKLIQSIGVSKAVDMAVKLGMRS
*** *.*.*:..*.**:************** .:** ::****::**:**
MSMEG_6201|M.smegmatis_MC2_155 YAQPGTARDYDPESSESLADFVKRQNIGSFTLGPIEVNALELSNVAATLA
TH_2122|M.thermoresistible__bu YAQPGTARDYDPNSDESIADWVKRQNIGSFTLGPIEVNALELSNVAATLA
Mflv_1365|M.gilvum_PYR-GCK YALPGTARDYDPESNESLADFVKRQNIGSFTLGPIEVNALELSNVAATLA
Mvan_5442|M.vanbaalenii_PYR-1 YALPGTARDYDPESNESLADFVKRQNLGSFTLGPIEVNALELSNVAATLA
Mb3707|M.bovis_AF2122/97 YANPGTARDYNPDSNESLADFVKRQNLGSFTLGPIELNALELSNVAATLA
Rv3682|M.tuberculosis_H37Rv YANPGTARDYNPDSNESLADFVKRQNLGSFTLGPIELNALELSNVAATLA
MMAR_5171|M.marinum_M YANPGTARDYNPDSNESLADFVKRQNIGSFTLGPIEVNALELSNVAATLA
MUL_4257|M.ulcerans_Agy99 YANPGTARDYNPDSNESLADFVKRQNIGSFTLGPIEVNALELSNVAATLA
MAV_0446|M.avium_104 YADPGTARDYNPDSNESLADFVKRQNIGSFTLGPIEVNALELSNVAATLA
MLBr_02308|M.leprae_Br4923 YTTPGTARDYNPDSNESLADFVKRQNIGSFTLGPIKVNALELSNVAATLA
MAB_0408c|M.abscessus_ATCC_199 YAEPGTAKPYDPEGDESLAEYIKKYNLGSFTLGPIQINPLELSNVAATLA
*: ****: *:*:..**:*:::*: *:********::*.***********
MSMEG_6201|M.smegmatis_MC2_155 SGGTWCPPNPIAQVVDRNGNEVAVTTETCEQVVPEGLANTLANAMSKDDT
TH_2122|M.thermoresistible__bu SGGVWCPPNPIDKVYDRHGNEVSVTTETCEQVVPEGLANTLANAMSEDDR
Mflv_1365|M.gilvum_PYR-GCK SGGMWCPPNPIAEVYDRYGEPVSVTTETCEQAVPEGLANTLANAMSKDDT
Mvan_5442|M.vanbaalenii_PYR-1 SGGMWCPPNPIAEVRDRYGELVSVTTETCEQVVPEGLANTLAVAMSKDDT
Mb3707|M.bovis_AF2122/97 SGGVWCPPNPIDQLIDRNGNEVAVTTETCDQVVPAGLANTLANAMSKDAV
Rv3682|M.tuberculosis_H37Rv SGGVWCPPNPIDQLIDRNGNEVAVTTETCDQVVPAGLANTLANAMSKDAV
MMAR_5171|M.marinum_M SGGVWCPPNPVDKLIDRNGNEVAVTTETCDQVVPEGLANTLANAMSKDAV
MUL_4257|M.ulcerans_Agy99 SGGVWCPPNPVDKLIDRNGNEVAVTTETCDQVVPEGLANTLANAMSKDAV
MAV_0446|M.avium_104 SGGTWCPPNPIDKLIDRKGNEVAVTTETCDQVVPEGLANTMANAMSKDAV
MLBr_02308|M.leprae_Br4923 SGGMWCPPNPIDKLFDRNGNEVAVTVETCDRVVPEGLANTLANAMSKDTK
MAB_0408c|M.abscessus_ATCC_199 SGGKWCPPNPIDKVFDRHGKEVPTTVEACEQVVPTGLANTLANALSKDDQ
*** ******: :: ** *: *..*.*:*::.** *****:* *:*:*
MSMEG_6201|M.smegmatis_MC2_155 GSGTAAASAGSVGWNLPMSGKTGTTEDNRSSAFLGFTNQYAAANYIYDDS
TH_2122|M.thermoresistible__bu GAGTAAGAAASVGWDLPMSGKTGTTESNRSSAFLGFTNQLAAANYIYDDS
Mflv_1365|M.gilvum_PYR-GCK GAGTAAGAAGSVGWNLPMSGKTGTTEANRSSAFLGFTNRYAAASYIYDDS
Mvan_5442|M.vanbaalenii_PYR-1 GAGTAAGAAGSTGWSLPMSGKTGTTESNRSSAFLGFTNQYAAASYIYDDS
Mb3707|M.bovis_AF2122/97 GSGTAAGSAGAAGWDLPMSGKTGTTEAHRSAGFVGFTNRYAAANYIYDDS
Rv3682|M.tuberculosis_H37Rv GSGTAAGSAGAAGWDLPMSGKTGTTEAHRSAGFVGFTNRYAAANYIYDDS
MMAR_5171|M.marinum_M GGGTASGSAGAAGWSLPVSGKTGTTEAHRSAGFVGFTNRYAAANYIYDDS
MUL_4257|M.ulcerans_Agy99 GGGTASGSAGAAGWSLPVSGKTGTTEAHRSAGFVGFTNRYAAANYIYDDS
MAV_0446|M.avium_104 GGGTAAGSAGAAGWDLPMSGKTGTTEAHRSSGFVGFTNHYAAANYIYDDS
MLBr_02308|M.leprae_Br4923 GSGTAAGSAGAAGWDLPMSGKTGTTEAHRSSGFVGFTNHYAAANYIYDDS
MAB_0408c|M.abscessus_ATCC_199 PGGTAAGSAGSVGWNLPLSSKTGTTEAHRSSAFLGYTNNLAGASYIYDDS
.***:.:*.:.**.**:*.****** :**:.*:*:**. *.*.******
MSMEG_6201|M.smegmatis_MC2_155 PNPSELCSFPLRQCGSGNLFGGNEPARTWFTAMKPIAESFGPVALPPTDP
TH_2122|M.thermoresistible__bu NSPGELCSFPLRRCGSGDLFGGNEPARTWFAAMKPIAEKFGPVALPPTDP
Mflv_1365|M.gilvum_PYR-GCK TAPTELCSFPLRQCGSGNLFGGNEPARTWFSAMQPIATAFGDVTLPPTDP
Mvan_5442|M.vanbaalenii_PYR-1 TAPTELCSFPLRQCGSGNLFGGNEPARTWFTAMTPIATAFGDVTLPPTDP
Mb3707|M.bovis_AF2122/97 SSPTDLCSGPLRHCGSGDLYGGNEPSRTWFAAMKPIANNFGEVQLPPTDP
Rv3682|M.tuberculosis_H37Rv SSPTDLCSGPLRHCGSGDLYGGNEPSRTWFAAMKPIANNFGEVQLPPTDP
MMAR_5171|M.marinum_M TNPTDLCSGPLRHCGNGDLYGGNEPARTWFTALKPIATDFGEVQLPPTDP
MUL_4257|M.ulcerans_Agy99 TNPTDLCSGPLRHCGNGDLYGGNEPARTWFTALKPIATDFGEVQLPPTDP
MAV_0446|M.avium_104 PNPTDLCSAPLRHCGEGDLYGGNEPARTWFTAMKPIATNFGPVQLPPTDP
MLBr_02308|M.leprae_Br4923 TSPTNLCSGPLRHCSNGDLYGGNEPARTWFTAMKPIATKFGEVRLPPTDP
MAB_0408c|M.abscessus_ATCC_199 TTPGDLCSFPLRKCGDGNLYGGNEPARAWFEAMKPIADNYGPTVLPPTEP
* :*** ***:*..*:*:*****:*:** *: *** :* . ****:*
MSMEG_6201|M.smegmatis_MC2_155 RYVDGGPGSRVPSVSGLTEEAARQRLKEAGFQVADQASYVNSGARSGTVV
TH_2122|M.thermoresistible__bu RYVDGGPGSRVPSVSGLSEEAARQRLRDAGFQVADGVATVNSSARRGTVV
Mflv_1365|M.gilvum_PYR-GCK RYVEGAPGSKVPSVAGMDQDTARARLREAGFQVADQATPVNSSSRYGTVV
Mvan_5442|M.vanbaalenii_PYR-1 RYVDGAPGSKVPSVAGMNQDTARQRLREAGFQVADQPTPVNSTAPYGAVV
Mb3707|M.bovis_AF2122/97 RYVDGAPGSRVPSVAGLDVDAARQRLKDAGFQVADQTNSVNSSAKYGEVV
Rv3682|M.tuberculosis_H37Rv RYVDGAPGSRVPSVAGLDVDAARQRLKDAGFQVADQTNSVNSSAKYGEVV
MMAR_5171|M.marinum_M RYVDGAPGSRVPSVAGLDVDQARQRLKDAGFQVADQPNAVNSSARYGEVV
MUL_4257|M.ulcerans_Agy99 RYVDGAPGSRVPSVAGLDVDQARQRLKDAGFQVADQPNAVNSSARYGEAV
MAV_0446|M.avium_104 RYVDGAPGSRVPSVAGLDIDAARQRIKEAGFQVADQNNFVNSSAKQGEVV
MLBr_02308|M.leprae_Br4923 RYVDGSPGSQVPSVTGLDLDAARQRLKGAGFQVADQPTLINSTAKLGAVV
MAB_0408c|M.abscessus_ATCC_199 RYVEGGPGSKVPSINGLREPEARDVLKKAGFRVADQASSVNSTAPFNEVV
***:*.***:***: *: ** :: ***:*** :** : . .*
MSMEG_6201|M.smegmatis_MC2_155 GTTPSGQTIPGSIITIQLSNGIPPPPPPPVN--IPLPGGP----------
TH_2122|M.thermoresistible__bu GTNPSGQTIPXXYPTQFLRADLDALGVIPADKLLEVPDGTRILIAGAVTH
Mflv_1365|M.gilvum_PYR-GCK GTAPSGQTVPGSIVTIQISNGIAPAPPPPPAG-APPPGGPP---------
Mvan_5442|M.vanbaalenii_PYR-1 GTSPTGQTVPGSIITIQISNGIAPPPPPPPPG-APP-GLPP---------
Mb3707|M.bovis_AF2122/97 GTSPSGQTIPGSIVTIQISNGIPPAPPPPPLPEDGGPPPP----------
Rv3682|M.tuberculosis_H37Rv GTSPSGQTIPGSIVTIQISNGIPPAPPPPPLPEDGGPPPP----------
MMAR_5171|M.marinum_M GTSPSGQTIPGSIITIQISNGIPPAPPPPP---EGEPPPP----------
MUL_4257|M.ulcerans_Agy99 GTSPSGQTIPGSIITIQISNGIPPAPPPPP---EGEPPPP----------
MAV_0446|M.avium_104 GTTPSGATIPGSIITIQVSNGIPPAPPPPP----EGAPPP----------
MLBr_02308|M.leprae_Br4923 GTTPSGQTIPGSIITIQTSSGIPPAPPPPP---EGGPPMPP---------
MAB_0408c|M.abscessus_ATCC_199 GTSPSGTTIPGSVITILMSNGIPPAPPPLVIPGIPGAPPGE---------
** *:* *:* * .: .
MSMEG_6201|M.smegmatis_MC2_155 ---PEIGQTVVEIPGLPPIT--VPVLGPP---------PPPPPP------
TH_2122|M.thermoresistible__bu RQRPATAQGVTFLN-LEDETGMVNVLCSPGVWARHRRLAQTAPALLVRGI
Mflv_1365|M.gilvum_PYR-GCK ---PPAGETVIQIPGLPPIT--VPVLGPP---------PPPPPPPG----
Mvan_5442|M.vanbaalenii_PYR-1 ---PPVGQTVIQIPGLPPIT--VPVLGPP---------PPPPPPPG----
Mb3707|M.bovis_AF2122/97 -----VGSQVVEIPGLPPIT--IPLLAPP----------PPAP--PP---
Rv3682|M.tuberculosis_H37Rv -----VGSQVVEIPGLPPIT--IPLLAPP----------PPAP--PP---
MMAR_5171|M.marinum_M -----VGSQVVEIPGLPPIT--IPLLAPP----------PP---------
MUL_4257|M.ulcerans_Agy99 -----VGSQVVEIPGLPPIT--IPLLAPP----------PP---------
MAV_0446|M.avium_104 -----VGSQVVEIPGLPPIT--IPLLAPP----------PPAPGQPPP--
MLBr_02308|M.leprae_Br4923 ----SVGSQVIEIPGLPPIT--IPLLAPP---------------------
MAB_0408c|M.abscessus_ATCC_199 -----MTTTIVNIPGLPPIT--VPVLAPP-------------P-------
: : * * : :* .*
MSMEG_6201|M.smegmatis_MC2_155 ------------------------------
TH_2122|M.thermoresistible__bu VQNATGAVTVVADRMRRLELRVGSRSRDFR
Mflv_1365|M.gilvum_PYR-GCK ------------------------------
Mvan_5442|M.vanbaalenii_PYR-1 ------------------------------
Mb3707|M.bovis_AF2122/97 ------------------------------
Rv3682|M.tuberculosis_H37Rv ------------------------------
MMAR_5171|M.marinum_M ------------------------------
MUL_4257|M.ulcerans_Agy99 ------------------------------
MAV_0446|M.avium_104 ------------------------------
MLBr_02308|M.leprae_Br4923 ------------------------------
MAB_0408c|M.abscessus_ATCC_199 ------------------------------