For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSADETRAIDILLVEDDPGDELITREAFEHNKIKNNLHVAHDGEEGLDFLYRRGPYADAPRPDLILLDLN LPKYDGRQLLEKIKSDEKLCHIPIVVLTTSSAEEDILRSYKLHANAYVTKPVDLDQFMSAVRQIDEFFVQ VVRLPQF*
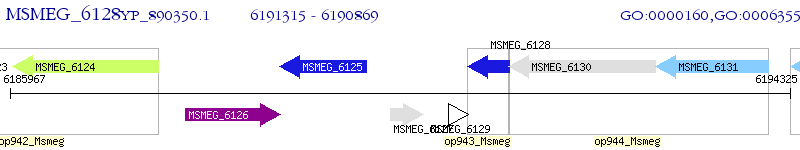
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6128 | - | - | 100% (148) | two-component system response regulator |
| M. smegmatis MC2 155 | MSMEG_5488 | - | 1e-08 | 26.77% (127) | DNA-binding response regulator |
| M. smegmatis MC2 155 | MSMEG_3246 | - | 7e-08 | 27.78% (144) | response regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1062c | trcR | 9e-10 | 27.42% (124) | two component transcriptional regulator TRCR |
| M. gilvum PYR-GCK | Mflv_3270 | - | 4e-72 | 89.19% (148) | response regulator receiver protein |
| M. tuberculosis H37Rv | Rv1033c | trcR | 9e-10 | 27.42% (124) | two component transcriptional regulator TRCR |
| M. leprae Br4923 | MLBr_00174 | - | 7e-09 | 26.19% (126) | putative two-component response regulator |
| M. abscessus ATCC 19977 | MAB_1926 | - | 3e-08 | 27.42% (124) | putative transcriptional regulatory protein PrrA |
| M. marinum M | MMAR_1265 | - | 3e-68 | 87.14% (140) | two-component transcriptional regulator |
| M. avium 104 | MAV_4240 | - | 3e-69 | 87.67% (146) | two-component system response regulator |
| M. thermoresistible (build 8) | TH_2174 | - | 3e-70 | 88.36% (146) | two-component system response regulator |
| M. ulcerans Agy99 | MUL_2623 | - | 9e-68 | 86.43% (140) | two component transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_2989 | - | 8e-73 | 89.19% (148) | response regulator receiver protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3270|M.gilvum_PYR-GCK ----------------------MTPAER--AIDVLLIEDDPGDELITREA
Mvan_2989|M.vanbaalenii_PYR-1 ----------------------MTPAER--AIDVLLIEDDPGDELITREA
MSMEG_6128|M.smegmatis_MC2_155 ----------------------MTSADETRAIDILLVEDDPGDELITREA
TH_2174|M.thermoresistible__bu ----------------------MTPTQEGRAIDVLLVEDDPGDELITREA
MAV_4240|M.avium_104 ----------------------MTTAGR--AIDILLVEDDPGDELITREA
MMAR_1265|M.marinum_M ----------------------MTAPGR--PIDILLIEDDPGDELITREA
MUL_2623|M.ulcerans_Agy99 ----------------------MTAPGR--PIDILLIEDDPGDELITREA
Mb1062c|M.bovis_AF2122/97 MTTMSGYTRSQRPRQAILGQLPRIHRADGSPIRVLLVDDEPALTNLVKMA
Rv1033c|M.tuberculosis_H37Rv MTTMSGYTRSQRPRQAILGQLPRIHRADGSPIRVLLVDDEPALTNLVKMA
MLBr_00174|M.leprae_Br4923 -------------------------------MRILAVDDDRAVRESLRRS
MAB_1926|M.abscessus_ATCC_1997 -----------------------MTQETTARHKVLMVDDDPDVRNSVARG
:* ::*: .
Mflv_3270|M.gilvum_PYR-GCK FEHNKIKNNLHVAHDGEEGLDFLYQRGVYEGAPRPDLILLDLNLPKYDGR
Mvan_2989|M.vanbaalenii_PYR-1 FEHNKIKNTLHVAHDGEEGLDFLYRRGAYTDAPRPDLVLLDLNLPKYDGR
MSMEG_6128|M.smegmatis_MC2_155 FEHNKIKNNLHVAHDGEEGLDFLYRRGPYADAPRPDLILLDLNLPKYDGR
TH_2174|M.thermoresistible__bu FEHNKIKNTLHVTRDGEEGLDFLYRRGRFADAPRPDLILLDLNLPKYDGR
MAV_4240|M.avium_104 FEHNKLNNRLHVAHDGEEGLNYLYRRGEFADAPRPDLILLDLNLPKYDGR
MMAR_1265|M.marinum_M FEHNKVNNRLHVAHDGEEGLDYLYQRGKYQQARRPDLILLDLNLPKYDGR
MUL_2623|M.ulcerans_Agy99 FEHNKVNNRLHVAHDGEEGLDYLYQRGKYQQARRPDLILLDLNLPKYDGR
Mb1062c|M.bovis_AF2122/97 LHYEGWD--VEVAHDGQEAIAKFDKV-------GPDVLVLDIMLPDVDGL
Rv1033c|M.tuberculosis_H37Rv LHYEGWD--VEVAHDGQEAIAKFDKV-------GPDVLVLDIMLPDVDGL
MLBr_00174|M.leprae_Br4923 LSFNGYS--VELANDGVEALEMVARD-------RPDALVLDVMMPRLDGL
MAB_1926|M.abscessus_ATCC_1997 LRHSGFD--VRVAVDGRDALTQLAAD-------EPDALVLDVQMPELDGV
: .. . :.:: ** :.: . ** ::**: :* **
Mflv_3270|M.gilvum_PYR-GCK QLLEQIKSDADLSHIPVVVLTTSSAEEDILRSYKLHANAYVTKPVDLDQF
Mvan_2989|M.vanbaalenii_PYR-1 QLLEQIKSDPDLSHIPVVVLTTSSAEEDILRSYKLHANAYVTKPVDLDQF
MSMEG_6128|M.smegmatis_MC2_155 QLLEKIKSDEKLCHIPIVVLTTSSAEEDILRSYKLHANAYVTKPVDLDQF
TH_2174|M.thermoresistible__bu QLLEQIKSDPDLSLIPVVVLTTSSAEEDILRSYQLHANAYVTKPVDLDQF
MAV_4240|M.avium_104 QLLEKIKSDPDLAQIPVVVLTTSSAEEDILKSYKLHANAYVTKPVDLDQF
MMAR_1265|M.marinum_M QLLEKIKSDSELCRIPVVVLTTSSAEEDILRSYNLHANAYVTKPVDLDQF
MUL_2623|M.ulcerans_Agy99 QLLEKIKFDSELCRIPVVVLTTSSAEEDILRSYNLHANAYVTKPVDLDQF
Mb1062c|M.bovis_AF2122/97 EILRRVRESD--VYTPTLFLTARDSVMDRVTGLTSGADDYMTKPFSLEEL
Rv1033c|M.tuberculosis_H37Rv EILRRVRESD--VYTPTLFLTARDSVMDRVTGLTSGADDYMTKPFSLEEL
MLBr_00174|M.leprae_Br4923 EVCRQLRSTG--DDLPILVLTARDSVSERVAGLDAGADDYLPKPFALEEL
MAB_1926|M.abscessus_ATCC_1997 AVVTALRALG--NDIPICVLSARDTVNDRIAGLEAGADDYLTKPFDLGEL
: :: * .*:: .: : : . *: *:.**. * ::
Mflv_3270|M.gilvum_PYR-GCK MNAVR------------------QIDEFFVQVVRLPQF------------
Mvan_2989|M.vanbaalenii_PYR-1 MNAVR------------------QIDEFFVQVVRLPQF------------
MSMEG_6128|M.smegmatis_MC2_155 MSAVR------------------QIDEFFVQVVRLPQF------------
TH_2174|M.thermoresistible__bu MSAVR------------------QIDEFFVQVVRLPTA------------
MAV_4240|M.avium_104 MKAVR------------------QIDEFFVQVVRLPSA------------
MMAR_1265|M.marinum_M MTAVR------------------QIDEFFLQVVRLPQS------------
MUL_2623|M.ulcerans_Agy99 MTAVR------------------QIDEFFLQVVRLPQS------------
Mb1062c|M.bovis_AF2122/97 VARLRGLLRRS-SHLERPADEALRVGDLTLDGASREVTRDGTPISLSSTE
Rv1033c|M.tuberculosis_H37Rv VARLRGLLRRS-SHLERPADEALRVGDLTLDGASREVTRDGTPISLSSTE
MLBr_00174|M.leprae_Br4923 LARIRALLRRTKPDD-AAESVAMSFSDLTLDPVTREVARGQRWISLTRTE
MAB_1926|M.abscessus_ATCC_1997 VARLRALLRRRGAGSGGLADSAITVGQITVDVSRRLVFVNGERVELSKRE
: :* ..:: ::
Mflv_3270|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2989|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6128|M.smegmatis_MC2_155 --------------------------------------------------
TH_2174|M.thermoresistible__bu --------------------------------------------------
MAV_4240|M.avium_104 --------------------------------------------------
MMAR_1265|M.marinum_M --------------------------------------------------
MUL_2623|M.ulcerans_Agy99 --------------------------------------------------
Mb1062c|M.bovis_AF2122/97 FELLRFLMRNPRRALSRTEILDRVWNYDFAGRTSIVDLYISYLRKKIDSD
Rv1033c|M.tuberculosis_H37Rv FELLRFLMRNPRRALSRTEILDRVWNYDFAGRTSIVDLYISYLRKKIDSD
MLBr_00174|M.leprae_Br4923 FALLEMLIANPRRVLTRSRILEEVWGFDFPTSGNALEVYVGYLRRKTEAD
MAB_1926|M.abscessus_ATCC_1997 FDLLVVLAENTGVVLSRTRLLELVWGYDFDVETNVADVFVSYLRRKLEAG
Mflv_3270|M.gilvum_PYR-GCK ---------------------
Mvan_2989|M.vanbaalenii_PYR-1 ---------------------
MSMEG_6128|M.smegmatis_MC2_155 ---------------------
TH_2174|M.thermoresistible__bu ---------------------
MAV_4240|M.avium_104 ---------------------
MMAR_1265|M.marinum_M ---------------------
MUL_2623|M.ulcerans_Agy99 ---------------------
Mb1062c|M.bovis_AF2122/97 REP-MIHTVRGIGYMLRPPE-
Rv1033c|M.tuberculosis_H37Rv REP-MIHTVRGIGYMLRPPE-
MLBr_00174|M.leprae_Br4923 GESRLIHTVRGVGYVLRETPP
MAB_1926|M.abscessus_ATCC_1997 GAARVIHTVRGVGYVMREEP-