For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTAGRAIDILLVEDDPGDELITREAFEHNKLNNRLHVAHDGEEGLNYLYRRGEFADAPRPDLILLDLNLP KYDGRQLLEKIKSDPDLAQIPVVVLTTSSAEEDILKSYKLHANAYVTKPVDLDQFMKAVRQIDEFFVQVV RLPSA*
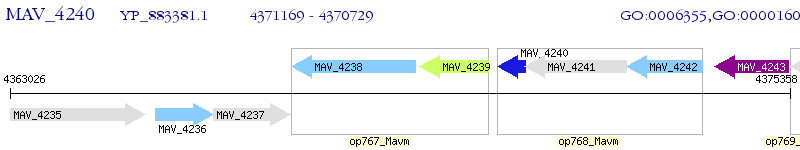
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4240 | - | - | 100% (146) | two-component system response regulator |
| M. avium 104 | MAV_1176 | - | 2e-09 | 27.56% (127) | two component transcriptional regulator trcr |
| M. avium 104 | MAV_1094 | - | 3e-09 | 26.19% (126) | two component response transcriptional regulatory protein MprA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1062c | trcR | 9e-10 | 27.56% (127) | two component transcriptional regulator TRCR |
| M. gilvum PYR-GCK | Mflv_3270 | - | 8e-69 | 86.11% (144) | response regulator receiver protein |
| M. tuberculosis H37Rv | Rv1033c | trcR | 9e-10 | 27.56% (127) | two component transcriptional regulator TRCR |
| M. leprae Br4923 | MLBr_00174 | - | 4e-09 | 26.19% (126) | putative two-component response regulator |
| M. abscessus ATCC 19977 | MAB_1076 | - | 8e-09 | 26.19% (126) | two component response transcriptional regulatory protein MprA |
| M. marinum M | MMAR_1265 | - | 7e-69 | 84.93% (146) | two-component transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_6128 | - | 3e-69 | 87.67% (146) | two-component system response regulator |
| M. thermoresistible (build 8) | TH_2174 | - | 1e-70 | 87.59% (145) | two-component system response regulator |
| M. ulcerans Agy99 | MUL_2623 | - | 2e-68 | 84.25% (146) | two component transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_2989 | - | 6e-71 | 87.50% (144) | response regulator receiver protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3270|M.gilvum_PYR-GCK ----------------------MTPAER--AIDVLLIEDDPGDELITREA
Mvan_2989|M.vanbaalenii_PYR-1 ----------------------MTPAER--AIDVLLIEDDPGDELITREA
MSMEG_6128|M.smegmatis_MC2_155 ----------------------MTSADETRAIDILLVEDDPGDELITREA
TH_2174|M.thermoresistible__bu ----------------------MTPTQEGRAIDVLLVEDDPGDELITREA
MAV_4240|M.avium_104 ----------------------MTTAGR--AIDILLVEDDPGDELITREA
MMAR_1265|M.marinum_M ----------------------MTAPGR--PIDILLIEDDPGDELITREA
MUL_2623|M.ulcerans_Agy99 ----------------------MTAPGR--PIDILLIEDDPGDELITREA
Mb1062c|M.bovis_AF2122/97 MTTMSGYTRSQRPRQAILGQLPRIHRADGSPIRVLLVDDEPALTNLVKMA
Rv1033c|M.tuberculosis_H37Rv MTTMSGYTRSQRPRQAILGQLPRIHRADGSPIRVLLVDDEPALTNLVKMA
MLBr_00174|M.leprae_Br4923 -------------------------------MRILAVDDDRAVRESLRRS
MAB_1076|M.abscessus_ATCC_1997 -----------------------------MPVRILVVDDDRAVRESLRRS
: :* ::*: . : :
Mflv_3270|M.gilvum_PYR-GCK FEHNKIKNNLHVAHDGEEGLDFLYQRGVYEGAPRPDLILLDLNLPKYDGR
Mvan_2989|M.vanbaalenii_PYR-1 FEHNKIKNTLHVAHDGEEGLDFLYRRGAYTDAPRPDLVLLDLNLPKYDGR
MSMEG_6128|M.smegmatis_MC2_155 FEHNKIKNNLHVAHDGEEGLDFLYRRGPYADAPRPDLILLDLNLPKYDGR
TH_2174|M.thermoresistible__bu FEHNKIKNTLHVTRDGEEGLDFLYRRGRFADAPRPDLILLDLNLPKYDGR
MAV_4240|M.avium_104 FEHNKLNNRLHVAHDGEEGLNYLYRRGEFADAPRPDLILLDLNLPKYDGR
MMAR_1265|M.marinum_M FEHNKVNNRLHVAHDGEEGLDYLYQRGKYQQARRPDLILLDLNLPKYDGR
MUL_2623|M.ulcerans_Agy99 FEHNKVNNRLHVAHDGEEGLDYLYQRGKYQQARRPDLILLDLNLPKYDGR
Mb1062c|M.bovis_AF2122/97 LHYEGWD--VEVAHDGQEAIAKFDKV-------GPDVLVLDIMLPDVDGL
Rv1033c|M.tuberculosis_H37Rv LHYEGWD--VEVAHDGQEAIAKFDKV-------GPDVLVLDIMLPDVDGL
MLBr_00174|M.leprae_Br4923 LSFNGYS--VELANDGVEALEMVARD-------RPDALVLDVMMPRLDGL
MAB_1076|M.abscessus_ATCC_1997 LSFNGYS--VDLAEDGVDALNVIAND-------RPDALVLDVMMPKLDGL
: .: . :.::.** :.: . . ** ::**: :* **
Mflv_3270|M.gilvum_PYR-GCK QLLEQIKSDADLSHIPVVVLTTSSAEEDILRSYKLHANAYVTKPVDLDQF
Mvan_2989|M.vanbaalenii_PYR-1 QLLEQIKSDPDLSHIPVVVLTTSSAEEDILRSYKLHANAYVTKPVDLDQF
MSMEG_6128|M.smegmatis_MC2_155 QLLEKIKSDEKLCHIPIVVLTTSSAEEDILRSYKLHANAYVTKPVDLDQF
TH_2174|M.thermoresistible__bu QLLEQIKSDPDLSLIPVVVLTTSSAEEDILRSYQLHANAYVTKPVDLDQF
MAV_4240|M.avium_104 QLLEKIKSDPDLAQIPVVVLTTSSAEEDILKSYKLHANAYVTKPVDLDQF
MMAR_1265|M.marinum_M QLLEKIKSDSELCRIPVVVLTTSSAEEDILRSYNLHANAYVTKPVDLDQF
MUL_2623|M.ulcerans_Agy99 QLLEKIKFDSELCRIPVVVLTTSSAEEDILRSYNLHANAYVTKPVDLDQF
Mb1062c|M.bovis_AF2122/97 EILRRVRESD--VYTPTLFLTARDSVMDRVTGLTSGADDYMTKPFSLEEL
Rv1033c|M.tuberculosis_H37Rv EILRRVRESD--VYTPTLFLTARDSVMDRVTGLTSGADDYMTKPFSLEEL
MLBr_00174|M.leprae_Br4923 EVCRQLRSTG--DDLPILVLTARDSVSERVAGLDAGADDYLPKPFALEEL
MAB_1076|M.abscessus_ATCC_1997 EVCRRLRSTG--DDLPILVLTARDSVSERVAGLDAGADDYLPKPFALEEL
:: .::: * :.**: .: : : . *: *:.**. *:::
Mflv_3270|M.gilvum_PYR-GCK MNAVR----------------------QIDEFFVQVVR------LPQF--
Mvan_2989|M.vanbaalenii_PYR-1 MNAVR----------------------QIDEFFVQVVR------LPQF--
MSMEG_6128|M.smegmatis_MC2_155 MSAVR----------------------QIDEFFVQVVR------LPQF--
TH_2174|M.thermoresistible__bu MSAVR----------------------QIDEFFVQVVR------LPTA--
MAV_4240|M.avium_104 MKAVR----------------------QIDEFFVQVVR------LPSA--
MMAR_1265|M.marinum_M MTAVR----------------------QIDEFFLQVVR------LPQS--
MUL_2623|M.ulcerans_Agy99 MTAVR----------------------QIDEFFLQVVR------LPQS--
Mb1062c|M.bovis_AF2122/97 VARLRGLLRRSSHLERPADEALRVGDLTLDGASREVTRDGTPISLSSTEF
Rv1033c|M.tuberculosis_H37Rv VARLRGLLRRSSHLERPADEALRVGDLTLDGASREVTRDGTPISLSSTEF
MLBr_00174|M.leprae_Br4923 LARIRALLRRTKPDDAAESVAMSFSDLTLDPVTREVARGQRWISLTRTEF
MAB_1076|M.abscessus_ATCC_1997 LARMRALLRRTTIDESADAATLSFGDLTLDPVTREVHRGTRSISLTRTEF
: :* :* :* * *.
Mflv_3270|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2989|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6128|M.smegmatis_MC2_155 --------------------------------------------------
TH_2174|M.thermoresistible__bu --------------------------------------------------
MAV_4240|M.avium_104 --------------------------------------------------
MMAR_1265|M.marinum_M --------------------------------------------------
MUL_2623|M.ulcerans_Agy99 --------------------------------------------------
Mb1062c|M.bovis_AF2122/97 ELLRFLMRNPRRALSRTEILDRVWNYDFAGRTSIVDLYISYLRKKIDSDR
Rv1033c|M.tuberculosis_H37Rv ELLRFLMRNPRRALSRTEILDRVWNYDFAGRTSIVDLYISYLRKKIDSDR
MLBr_00174|M.leprae_Br4923 ALLEMLIANPRRVLTRSRILEEVWGFDFPTSGNALEVYVGYLRRKTEADG
MAB_1076|M.abscessus_ATCC_1997 ALLEMLMANPRRVLTRSRILEEVWGFDFPTSGNALEVYVGYLRRKTEAEG
Mflv_3270|M.gilvum_PYR-GCK --------------------
Mvan_2989|M.vanbaalenii_PYR-1 --------------------
MSMEG_6128|M.smegmatis_MC2_155 --------------------
TH_2174|M.thermoresistible__bu --------------------
MAV_4240|M.avium_104 --------------------
MMAR_1265|M.marinum_M --------------------
MUL_2623|M.ulcerans_Agy99 --------------------
Mb1062c|M.bovis_AF2122/97 E-PMIHTVRGIGYMLRPPE-
Rv1033c|M.tuberculosis_H37Rv E-PMIHTVRGIGYMLRPPE-
MLBr_00174|M.leprae_Br4923 ESRLIHTVRGVGYVLRETPP
MAB_1076|M.abscessus_ATCC_1997 ELRLIHTVRGVGYVLRETPP