For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTGSTTDADRPRRVLIAEDEALIRLDLAEMLREEGYEVVGEAGDGQEAVEMAESLRPDLVIMDVKMPRRD GIDAASEIASKRIAPIVILTAFSQRELVERARDAGAMAYLVKPFNINDLVPAIEVAVSRFAELSALETEV ATLSERLETRKLVERAKGLLQAKHKMTEPEAFKWIQRAAMDRRTTMKRVAEVVLETLDDTKQAPAPEQ
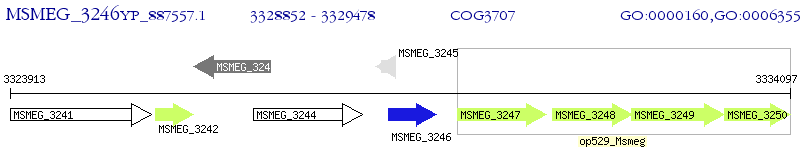
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3246 | - | - | 100% (208) | response regulator |
| M. smegmatis MC2 155 | MSMEG_5662 | prrA | 9e-14 | 32.12% (137) | DNA-binding response regulator PrrA |
| M. smegmatis MC2 155 | MSMEG_1874 | mtrA | 9e-14 | 33.05% (118) | DNA-binding response regulator MtrA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1652 | - | 6e-94 | 87.19% (203) | two-component system transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_3575 | - | 6e-94 | 88.00% (200) | response regulator receiver/ANTAR domain-containing protein |
| M. tuberculosis H37Rv | Rv1626 | - | 6e-94 | 87.19% (203) | two-component system transcriptional regulator |
| M. leprae Br4923 | MLBr_01286 | - | 1e-91 | 84.73% (203) | putative two-component response regulatory protein |
| M. abscessus ATCC 19977 | MAB_2627c | - | 1e-86 | 82.50% (200) | two-component response regulatory protein |
| M. marinum M | MMAR_2429 | - | 2e-93 | 85.85% (205) | two-component system transcriptional regulator |
| M. avium 104 | MAV_3158 | - | 6e-93 | 86.27% (204) | response regulator |
| M. thermoresistible (build 8) | TH_1079 | - | 1e-90 | 87.56% (193) | Probable two-component system transcriptional regulator |
| M. ulcerans Agy99 | MUL_1608 | - | 1e-93 | 85.85% (205) | two-component system transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_2840 | - | 1e-92 | 88.72% (195) | response regulator receiver/ANTAR domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3246|M.smegmatis_MC2_155 ----------------MTGSTTD----ADRPRRVLIAEDEALIRLDLAEM
TH_1079|M.thermoresistible__bu ---------------------------------VLVAEDEALIRLDLAEM
Mflv_3575|M.gilvum_PYR-GCK MAPRSPISPAGFTMFSMSSASASPSDAAATPHRVLIAEDEALIRMDLAEM
Mvan_2840|M.vanbaalenii_PYR-1 ----------------MSSASASPSDAAVTPRRVLVAEDEALIRLDLAEM
MMAR_2429|M.marinum_M ----------------MTGPTTDAD--AAKTRRVLIAEDEALIRMDLAEM
MUL_1608|M.ulcerans_Agy99 -------------MSVMTGPTTDAD--AAKTRRVLIAEDEALIRMDLAEM
Mb1652|M.bovis_AF2122/97 ----------------MTGPTTDAD--AAVPRRVLIAEDEALIRMDLAEM
Rv1626|M.tuberculosis_H37Rv ----------------MTGPTTDAD--AAVPRRVLIAEDEALIRMDLAEM
MAV_3158|M.avium_104 -------------MSVMTGPTTDTD--AAVPRRVLIAEDEALIRLDLAEM
MLBr_01286|M.leprae_Br4923 ----------------MTGPTTDAD--AATPHRVLIAEDEALIRLDLAEM
MAB_2627c|M.abscessus_ATCC_199 ----------------MTGPQNTEP--AAPARRVLVAEDEALIRLDLVEM
**:********:**.**
MSMEG_3246|M.smegmatis_MC2_155 LREEGYEVVGEAGDGQEAVEMAESLRPDLVIMDVKMPRRDGIDAASEIAS
TH_1079|M.thermoresistible__bu LRDEGYDVVGEAGDGQEAVELAESLRPDLVIMDVKMPRRDGIDAAREIAA
Mflv_3575|M.gilvum_PYR-GCK LREEGYEIVGEAGDGQEAVDLAESLRPDLVIMDVKMPRRDGIDAASEIAS
Mvan_2840|M.vanbaalenii_PYR-1 LREEGYEIVGEAGDGQEAVELAEALRPDLVIMDVKMPRRDGIDAASEIAS
MMAR_2429|M.marinum_M LREEGYDIVGEAGDGQEAVELAELHKPDLVIMDVKMPRRDGIDAASEIAT
MUL_1608|M.ulcerans_Agy99 LREEGYDIVGEAGDGQEAVELAELHKPDLVIMDVKMPRRDGIDAASEIAT
Mb1652|M.bovis_AF2122/97 LREEGYEIVGEAGDGQEAVELAELHKPDLVIMDVKMPRRDGIDAASEIAS
Rv1626|M.tuberculosis_H37Rv LREEGYEIVGEAGDGQEAVELAELHKPDLVIMDVKMPRRDGIDAASEIAS
MAV_3158|M.avium_104 LREEGYEIVGEAGDGQEAVELAERHRPDLVIMDVKMPRRDGIDAASEIAS
MLBr_01286|M.leprae_Br4923 LRDEGYDIVGEAADGQEAVELAERHKPDLVIMDVKMPRRDGIDAASEIAS
MAB_2627c|M.abscessus_ATCC_199 LHEEGYDVVGQAGDGQEAVDLAEELKPDLVIMDVKMPRRDGIDAASEIAT
*::***::**:*.******::** :******************* ***:
MSMEG_3246|M.smegmatis_MC2_155 KRIAPIVILTAFSQRELVERARDAGAMAYLVKPFNINDLVPAIEVAVSRF
TH_1079|M.thermoresistible__bu KRIAPVVVLTAFSQRDLVEQARDAGAMAYLVKPFSITDLIPAIEVAVSRF
Mflv_3575|M.gilvum_PYR-GCK KRIAPIVILTAFSQRELVERARDAGAMAYLVKPFSITDLIPAIEVAVSRF
Mvan_2840|M.vanbaalenii_PYR-1 KRIAPIVILTAFSQRELVERARDAGAMAYLVKPFSITDLIPAIEVAVSRF
MMAR_2429|M.marinum_M KRIAPIVVLTAFSQRDLVERARDAGAMAYLVKPFTVSDLVPAIELAVSRF
MUL_1608|M.ulcerans_Agy99 KRIAPIVVLTAFSQRDLVERARDAGAMAYLVKPFTVSDLVPAIELAVSRF
Mb1652|M.bovis_AF2122/97 KRIAPIVVLTAFSQRDLVERARDAGAMAYLVKPFSISDLIPAIELAVSRF
Rv1626|M.tuberculosis_H37Rv KRIAPIVVLTAFSQRDLVERARDAGAMAYLVKPFSISDLIPAIELAVSRF
MAV_3158|M.avium_104 KRIAPIVVLTAFSQRDLVERARDAGAMAYLVKPFTISDLIPAIELAVSRF
MLBr_01286|M.leprae_Br4923 KRIAPIVVLTAFSQRDLVERARDAGAMAYLVKPFTISDLIPAIALAMSRF
MAB_2627c|M.abscessus_ATCC_199 KRIAPIVILTAFSQRDLVEKARDAGAMAYLVKPFSKTDLVPAIEVAVSRF
*****:*:*******:***:**************. .**:*** :*:***
MSMEG_3246|M.smegmatis_MC2_155 AELSALETEVATLSERLETRKLVERAKGLLQAKHKMTEPEAFKWIQRAAM
TH_1079|M.thermoresistible__bu AEITALESEVAELSERLETRKLVERAKGLLQAKHNMTEPEAFKWIQRAAM
Mflv_3575|M.gilvum_PYR-GCK SEIAELEKEVATLSDRLETRKLVERAKGLLQAKQGMTEPEAFKWIQRAAM
Mvan_2840|M.vanbaalenii_PYR-1 SEMTELEKEVANLSDRLETRKLVERAKGLLQAKQGMSEPEAFKWIQRAAM
MMAR_2429|M.marinum_M SEISALENEVATLSDRLETRKLVERAKGLLQAKQGMTEPEAFKWIQRAAM
MUL_1608|M.ulcerans_Agy99 SEISALENEVATLSDRLETRKLVERAKGLLQAKQGMTEPEAFKWIQRAAM
Mb1652|M.bovis_AF2122/97 REITALEGEVATLSERLETRKLVERAKGLLQTKHGMTEPDAFKWIQRAAM
Rv1626|M.tuberculosis_H37Rv REITALEGEVATLSERLETRKLVERAKGLLQTKHGMTEPDAFKWIQRAAM
MAV_3158|M.avium_104 SEIAELEREVATLSERLETRKLVERAKGLLQTEQGMTEPEAFKWIQRAAM
MLBr_01286|M.leprae_Br4923 SELTALEREVATLADRLETRKLVERAKGLLQVKQGMTEPEAFKWIQRAAM
MAB_2627c|M.abscessus_ATCC_199 GELTALEKEVQTLADRLETRKLVERAKGVLQSTQGLTEPEAFKWIQRAAM
*:: ** ** *::*************:** : ::**:**********
MSMEG_3246|M.smegmatis_MC2_155 DRRTTMKRVAEVVLETLDDTKQAPAPEQ-
TH_1079|M.thermoresistible__bu DRRTTMKRVAEVVLENLDTPKDQTPSN--
Mflv_3575|M.gilvum_PYR-GCK DRRTTMKRVAEVVLETLDPPAASPEPEAT
Mvan_2840|M.vanbaalenii_PYR-1 DRRTTMKRVAEVVLETLDPPADTPAGPGQ
MMAR_2429|M.marinum_M DRRTTMKRVAEVVLETLDTPDEA------
MUL_1608|M.ulcerans_Agy99 DRRTTMKRVAEVVLETLDTPDEA------
Mb1652|M.bovis_AF2122/97 DRRTTMKRVAEVVLETLGTPKDT------
Rv1626|M.tuberculosis_H37Rv DRRTTMKRVAEVVLETLGTPKDT------
MAV_3158|M.avium_104 DRRTTMKRVAEVVLETLGTPKE-------
MLBr_01286|M.leprae_Br4923 DRRTTMKRVAEVVLETLDTPKDT------
MAB_2627c|M.abscessus_ATCC_199 DRRTTMKRVAEVVLETLDTDAPS------
***************.*.