For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
HVLVADAHLLPHRSRLEAALPAGVTVCWREPGDSRILDDLPEADVYVGSRFTADMAAVAHRLRLIHVNGA GTDKIDFAHLRADTLVANTFHHEQSIAEYIAGAAVLLRRDFLAQDRALRTGRWATPGYDRQIPQPRTLRD ARIGYIGFGHIGRTSWKLLRAFGADAVAVTGTGRVDAAAEGLRWAGDTGRLTALMTESDVVVVCAPLTER TVGMIGADELRALGPSGVLINVGRGPLVVESALYEALSAGTIAAAALDVWYRYPGPDGDGTPSDLPFDTL PNVLMTPHVSGVTADTFAARADDIAANIGRLQRGEPLVNQIDRMDRQSGHQLS*
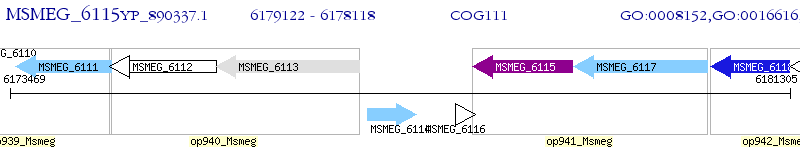
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6115 | - | - | 100% (334) | phosphoglycerate dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0604 | - | 8e-32 | 33.33% (327) | glyoxylate reductase |
| M. smegmatis MC2 155 | MSMEG_2496 | - | 1e-28 | 32.49% (277) | NAD-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3020c | serA1 | 2e-22 | 28.85% (305) | D-3-phosphoglycerate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0334 | - | 1e-30 | 33.02% (324) | D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding |
| M. tuberculosis H37Rv | Rv2996c | serA1 | 2e-22 | 28.85% (305) | D-3-phosphoglycerate dehydrogenase |
| M. leprae Br4923 | MLBr_01692 | serA | 1e-21 | 27.63% (304) | D-3-phosphoglycerate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4334 | - | 1e-31 | 34.23% (333) | 2-hydroxyacid dehydrogenase |
| M. marinum M | MMAR_5282 | serA4 | 6e-27 | 31.61% (310) | D-3-phosphoglycerate dehydrogenase SerA4 |
| M. avium 104 | MAV_0349 | - | 2e-24 | 30.75% (322) | D-isomer specific 2-hydroxyacid dehydrogenase family protein |
| M. thermoresistible (build 8) | TH_3383 | - | 4e-27 | 31.90% (326) | PUTATIVE D-isomer specific 2-hydroxyacid dehydrogenase, |
| M. ulcerans Agy99 | MUL_4355 | serA4 | 2e-25 | 30.32% (310) | D-3-phosphoglycerate dehydrogenase SerA4 |
| M. vanbaalenii PYR-1 | Mvan_1016 | - | 1e-114 | 64.40% (323) | D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5282|M.marinum_M ----------------MSQPASHGFGQDHVPRVAILDDYARVATDLADWS
MUL_4355|M.ulcerans_Agy99 ----------------MSQPASHGFGQDHVPRVAILDDYARVATDLADWS
MAV_0349|M.avium_104 -----------------------------MPRLAILDDYARVALQVADWS
Mb3020c|M.bovis_AF2122/97 --------------------------------MSLPVVLIADKLAPSTVA
Rv2996c|M.tuberculosis_H37Rv --------------------------------MSLPVVLIADKLAPSTVA
MLBr_01692|M.leprae_Br4923 --------------------------------MDLPVVLIADKLAQSTVA
Mflv_0334|M.gilvum_PYR-GCK --------------------------------MRVLAHFLPGPKVTEFLS
TH_3383|M.thermoresistible__bu VDRPAVALSANRRRHCPGMLRGDMSGVNSRSALRVLAHFRPGPKVLEFLE
MAB_4334|M.abscessus_ATCC_1997 -----------------------MSLSGENHRVRVLAHFPSGPRVLEQLA
MSMEG_6115|M.smegmatis_MC2_155 -----------------------------MHVLVADAHLLPHRSRLEAAL
Mvan_1016|M.vanbaalenii_PYR-1 -----------------------------MRVLVVDPNLVPHRERLEAAM
:
MMAR_5282|M.marinum_M PVRGRATLTVFDRHLGQAEAAEALADFDAVCTMRERMAFPRSLLERLPRL
MUL_4355|M.ulcerans_Agy99 PVRGRATLTVFDRHLGQAEAAEALADFDAVCTMRERMAFPRSLLERLPRL
MAV_0349|M.avium_104 RVQKRCDITVFDEHLSEQQAVDALRPFDVVCTMRERMAFPRSLIERLPHL
Mb3020c|M.bovis_AF2122/97 ALGDQVEVRWVDG--PDRDKLLAAVPEADALLVRSATTVDAEVLAAAPKL
Rv2996c|M.tuberculosis_H37Rv ALGDQVEVRWVDG--PDRDKLLAAVPEADALLVRSATTVDAEVLAAAPKL
MLBr_01692|M.leprae_Br4923 ALGDQVEVRWVDG--PDRTKLLAAVPEADALLVRSATTVDAEVLAAAPKL
Mflv_0334|M.gilvum_PYR-GCK PHTDWLDIRYCAE--DDDETFYRELPDAEVIWH-VLRPISGDDLSKAPKL
TH_3383|M.thermoresistible__bu PESGWLDVRFCDE--DDDETFHRELPHAEVIWH-VLRPLSGEDLTRATRC
MAB_4334|M.abscessus_ATCC_1997 PHADWLDVRFCAE--DDDNTFYAELPGADVLWH-VLRPLSADDVAKGERL
MSMEG_6115|M.smegmatis_MC2_155 PAG--VTVCWREP---GDSRILDDLPEADVYVG---SRFTADMAAVAHRL
Mvan_1016|M.vanbaalenii_PYR-1 PPG--TEVLWHVA---GVP--EDELRKAEVFVG---SRFTAEMARLAENL
: . . . .
MMAR_5282|M.marinum_M KLITIVGMNLPNLDLAAATERGVLVAHSNFAHPRFRAARDATAEFAWGLL
MUL_4355|M.ulcerans_Agy99 KLITIVGMNLPNLDLAAATERGVLVAHSNFAHPRFRAARDATAEFAWGLL
MAV_0349|M.avium_104 RLLTIVGRQLPNLDMAAATEHGIAVARSEFAHPRFAALRDATPELAWGLM
Mb3020c|M.bovis_AF2122/97 KIVARAGVGLDNVDVDAATARGVLVVNAPTSN------IHSAAEHALALL
Rv2996c|M.tuberculosis_H37Rv KIVARAGVGLDNVDVDAATARGVLVVNAPTSN------IHSAAEHALALL
MLBr_01692|M.leprae_Br4923 KIVARAGVGLDNVDVDAATARGVLVVNAPTSN------IHSAAEHALALL
Mflv_0334|M.gilvum_PYR-GCK RLVHKLGAGVNTVDVDTATARGIAVANMPGAN------APSVAEGTLLLM
TH_3383|M.thermoresistible__bu RLVHKLGTGVNTIDVDTATERGIAVANMPGAN------AASVAEGTVMLM
MAB_4334|M.abscessus_ATCC_1997 RLIHKFGAGVNTIALDAAVEHGVAVANMPGAN------APSVAEGALLLM
MSMEG_6115|M.smegmatis_MC2_155 RLIHVNGAGTDKIDFAHLRADTLVANTFHHEQ--------SIAEYIAGAA
Mvan_1016|M.vanbaalenii_PYR-1 RLLHPAAAGTDKIDLSALSADVLVANTFHHER--------SIAEYVVGAA
::: . .: . : . . : .*
MMAR_5282|M.marinum_M IATVRNLAEEHRQLRAGGWQTTTGMT-------LSGKTLGLLGLGRVGKR
MUL_4355|M.ulcerans_Agy99 IATVHNLAEEHRQVRAGGWQTTTGMT-------LSGKTLGLLGLGRVGKR
MAV_0349|M.avium_104 IATVRHLADEHRQMRAGGWQRTAGMT-------LSGKTLGLLGLGRVGSR
Mb3020c|M.bovis_AF2122/97 LAASRQIPAADASLREHTWKRSSFSG-----TEIFGKTVGVVGLGRIGQL
Rv2996c|M.tuberculosis_H37Rv LAASRQIPAADASLREHTWKRSSFSG-----TEIFGKTVGVVGLGRIGQL
MLBr_01692|M.leprae_Br4923 LAASRQIAEADASLRAHIWKRSSFSG-----TEIFGKTVGVVGLGRIGQL
Mflv_0334|M.gilvum_PYR-GCK LAALRRLPELDRATRTGNGWPSDPTLGESV-RDLGGCTVGLVGYGNIAKQ
TH_3383|M.thermoresistible__bu LAALRRLVELDRATRAGTGWPTDPSLGETV-RDIGGSTVGLIGYGNVARR
MAB_4334|M.abscessus_ATCC_1997 LAALRQLPRLDRDIRSGNGWPTDQSLGESV-RDIGSCTIGLVGYGNIAKS
MSMEG_6115|M.smegmatis_MC2_155 VLLRRDFLAQDRALRTGRWATPGYDRQIPQPRTLRDARIGYIGFGHIGRT
Mvan_1016|M.vanbaalenii_PYR-1 VMLRRDFLTQDARLRDGVWASSVYDSTVPQPSTLADAHVGFVGFGHIGRH
: : : . * . : . :* :* *.:.
MMAR_5282|M.marinum_M MTEYAKAFGMDVIAWSQN---LTEEAAAAHGVRRVDKPVLFAQSDVVSIH
MUL_4355|M.ulcerans_Agy99 MTEYAKAFGMDVIAWSQN---LTEEAAAAHGVRRVDKPVLFAQSDMVSIH
MAV_0349|M.avium_104 MAGFAAAFGMEVIAWSQN---LTDDAAAAVGARRVSKAALFESADVVSVH
Mb3020c|M.bovis_AF2122/97 VAQRIAAFGAYVVAYDPY---VSPARAAQLGIELLSLDDLLARADFISVH
Rv2996c|M.tuberculosis_H37Rv VAQRIAAFGAYVVAYDPY---VSPARAAQLGIELLSLDDLLARADFISVH
MLBr_01692|M.leprae_Br4923 VAARIAAFGAHVIAYDPY---VAPARAAQLGIELMSFDDLLARADFISVH
Mflv_0334|M.gilvum_PYR-GCK VERILLAIGAEVVHTSTR---DDGLPGWRT------LPDLLSESDIVSLH
TH_3383|M.thermoresistible__bu VEQIVAAMGAEVLHTSTA---DDGHPNWRP------LNDLLARSDIVSIH
MAB_4334|M.abscessus_ATCC_1997 LEKILLAMGATVLHTSTR---DDGTAGWRG------LNDLLTSSDIVSLH
MSMEG_6115|M.smegmatis_MC2_155 SWKLLRAFGADAVAVTGTGRVDAAAEGLRWAGDTGRLTALMTESDVVVVC
Mvan_1016|M.vanbaalenii_PYR-1 TWKLLQVFGCTAAAVTGSGRVADDAG-LAWVDDTGKLERLMRECDVVVVS
.:* . *: .*.: :
MMAR_5282|M.marinum_M VVLSERTRALVGAGELAAMKPGAYLINTSRGPIVDEGALIGALEAGRIAG
MUL_4355|M.ulcerans_Agy99 VVLSERTRALVGAGELAAMKPGAYLINTSRGPIVDEGALIGALEAGRIAG
MAV_0349|M.avium_104 LVLSERTRGLVGEPELALMKPSAYLINTARGPIVDEPALIAALTAGRIAG
Mb3020c|M.bovis_AF2122/97 LPKTPETAGLIDKEALAKTKPGVIIVNAARGGLVDEAALADAITGGHVRA
Rv2996c|M.tuberculosis_H37Rv LPKTPETAGLIDKEALAKTKPGVIIVNAARGGLVDEAALADAITGGHVRA
MLBr_01692|M.leprae_Br4923 LPKTPETAGLIDKEALAKTKPGVIIVNAARGGLVDEVALADAVRSGHVRA
Mflv_0334|M.gilvum_PYR-GCK LPLTGATEGLLGRKALAQMKTDAVLVNTSRGAIVDEAALVDALRAGTLAA
TH_3383|M.thermoresistible__bu VPLTEKTLGLLDATALSRMKPAAVLVNTARGAIVDEIALTEALHARRLAA
MAB_4334|M.abscessus_ATCC_1997 LPLTEASTGMLDSAALNRMKPGSVLVNTSRGAVVDEAALVNALQQGPLGA
MSMEG_6115|M.smegmatis_MC2_155 APLTERTVGMIGADELRALGPSGVLINVGRGPLVVESALYEALSAGTIAA
Mvan_1016|M.vanbaalenii_PYR-1 APLNRHTEGMIGAAELKALGPDGVLINVGRGPLVQERALYDALAGGAIKA
. : .::. * . ::*..** :* * ** *: : .
MMAR_5282|M.marinum_M AGLDVYDVEPLPV------DHRLRSLPNVTLSPHLGYVTREMLSACYADT
MUL_4355|M.ulcerans_Agy99 AGLDVYDVEPLPV------DHRLRSLPNVTLSPHLGYVTREMLSVCYADT
MAV_0349|M.avium_104 AGLDVYDVEPLPG------DHPLRSLPNVTLSPHLGYVTREMLGAFYADT
Mb3020c|M.bovis_AF2122/97 AGLDVFATEPCT-------DSPLFELAQVVVTPHLGASTAEAQDRAGTDV
Rv2996c|M.tuberculosis_H37Rv AGLDVFATEPCT-------DSPLFELAQVVVTPHLGASTAEAQDRAGTDV
MLBr_01692|M.leprae_Br4923 AGLDVFATEPCT-------DSPLFELSQVVVTPHLGASTAEAQDRAGTDV
Mflv_0334|M.gilvum_PYR-GCK AGLDVFAVEPVPS------DNPLLGLPNVVLTPHVTWYTADTMRRYLEFA
TH_3383|M.thermoresistible__bu AGLDVFATEPVRP------NNPLLTLDNVVLTPHVSWYTVDTMRRYLTHA
MAB_4334|M.abscessus_ATCC_1997 AGLDVFAQEPVSP------ENPLLALPNVVLTPHVTWFTADTMTRYLHHA
MSMEG_6115|M.smegmatis_MC2_155 AALDVWYRYPGPDGDGTPSDLPFDTLPNVLMTPHVSGVTADTFAARADDI
Mvan_1016|M.vanbaalenii_PYR-1 AAVDVWYRYPADGTGCAPSDLPFATLPGILMTPHTSGVTTDTFKGRVDDV
*.:**: * : : * : ::** * :
MMAR_5282|M.marinum_M VEAVVAWLDGAPIRIANPQAVQRPPS------------------------
MUL_4355|M.ulcerans_Agy99 VEAVVAWLDGAPIRIANPQAVRRPPS------------------------
MAV_0349|M.avium_104 VESVLAWLDGTPIRIANPEVLQRD--------------------------
Mb3020c|M.bovis_AF2122/97 AESVRLALAGEFVPDAVNVGGGVVNEEVAPWLDLVRKLGVLAGVLSDELP
Rv2996c|M.tuberculosis_H37Rv AESVRLALAGEFVPDAVNVGGGVVNEEVAPWLDLVRKLGVLAGVLSDELP
MLBr_01692|M.leprae_Br4923 AESVRLALAGEFVPDAVNVDGGVVNEEVAPWLDLVCKLGVLVAALSDELP
Mflv_0334|M.gilvum_PYR-GCK LDNCERLRDGRALAHVVNDPSG----------------------------
TH_3383|M.thermoresistible__bu VDNCRRLRDGEQLADVVNGITGRRPG------------------------
MAB_4334|M.abscessus_ATCC_1997 IDNCRRIHEGMPLADRVR--------------------------------
MSMEG_6115|M.smegmatis_MC2_155 AANIGRLQRGEPLVNQIDRMDRQSGHQLS---------------------
Mvan_1016|M.vanbaalenii_PYR-1 AANVGRLQNGEPLRNLVPH-------------------------------
* :
MMAR_5282|M.marinum_M --------------------------------------------------
MUL_4355|M.ulcerans_Agy99 --------------------------------------------------
MAV_0349|M.avium_104 --------------------------------------------------
Mb3020c|M.bovis_AF2122/97 VSLSVQVRGELAAEEVEVLRLSALRGLFSAVIEDAVTFVNAPALAAERGV
Rv2996c|M.tuberculosis_H37Rv VSLSVQVRGELAAEEVEVLRLSALRGLFSAVIEDAVTFVNAPALAAERGV
MLBr_01692|M.leprae_Br4923 ASLSVHVRGELASEDVEILRLSALRGLFSTVIEDAVTFVNAPALAAERGV
Mflv_0334|M.gilvum_PYR-GCK --------------------------------------------------
TH_3383|M.thermoresistible__bu --------------------------------------------------
MAB_4334|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6115|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_1016|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_5282|M.marinum_M --------------------------------------------------
MUL_4355|M.ulcerans_Agy99 --------------------------------------------------
MAV_0349|M.avium_104 --------------------------------------------------
Mb3020c|M.bovis_AF2122/97 TAEICKASESPNHRSVVDVRAVGADGSVVTVSGTLYGPQLSQKIVQINGR
Rv2996c|M.tuberculosis_H37Rv TAEICKASESPNHRSVVDVRAVGADGSVVTVSGTLYGPQLSQKIVQINGR
MLBr_01692|M.leprae_Br4923 SAEITTGSESPNHRSVVDVRAVASDGSVVNIAGTLSGPQLVQKIVQVNGR
Mflv_0334|M.gilvum_PYR-GCK --------------------------------------------------
TH_3383|M.thermoresistible__bu --------------------------------------------------
MAB_4334|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6115|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_1016|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_5282|M.marinum_M --------------------------------------------------
MUL_4355|M.ulcerans_Agy99 --------------------------------------------------
MAV_0349|M.avium_104 --------------------------------------------------
Mb3020c|M.bovis_AF2122/97 HFDLRAQGINLIIHYVDRPGALGKIGTLLGTAGVNIQAAQLSEDAEGPGA
Rv2996c|M.tuberculosis_H37Rv HFDLRAQGINLIIHYVDRPGALGKIGTLLGTAGVNIQAAQLSEDAEGPGA
MLBr_01692|M.leprae_Br4923 NFDLRAQGMNLVIRYVDQPGALGKIGTLLGAAGVNIQAAQLSEDTEGPGA
Mflv_0334|M.gilvum_PYR-GCK --------------------------------------------------
TH_3383|M.thermoresistible__bu --------------------------------------------------
MAB_4334|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6115|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_1016|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_5282|M.marinum_M ---------------------------------
MUL_4355|M.ulcerans_Agy99 ---------------------------------
MAV_0349|M.avium_104 ---------------------------------
Mb3020c|M.bovis_AF2122/97 TILLRLDQDVPDDVRTAIAAAVDAYKLEVVDLS
Rv2996c|M.tuberculosis_H37Rv TILLRLDQDVPDDVRTAIAAAVDAYKLEVVDLS
MLBr_01692|M.leprae_Br4923 TILLRLDQDVPGDVRSAIVAAVSANKLEVVNLS
Mflv_0334|M.gilvum_PYR-GCK ---------------------------------
TH_3383|M.thermoresistible__bu ---------------------------------
MAB_4334|M.abscessus_ATCC_1997 ---------------------------------
MSMEG_6115|M.smegmatis_MC2_155 ---------------------------------
Mvan_1016|M.vanbaalenii_PYR-1 ---------------------------------