For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SQPASHGFGQDHVPRVAILDDYARVATDLADWSPVRGRATLTVFDRHLGQAEAAEALADFDAVCTMRERM AFPRSLLERLPRLKLITIVGMNLPNLDLAAATERGVLVAHSNFAHPRFRAARDATAEFAWGLLIATVHNL AEEHRQVRAGGWQTTTGMTLSGKTLGLLGLGRVGKRMTEYAKAFGMDVIAWSQNLTEEAAAAHGVRRVDK PVLFAQSDMVSIHVVLSERTRALVGAGELAAMKPGAYLINTSRGPIVDEGALIGALEAGRIAGAGLDVYD VEPLPVDHRLRSLPNVTLSPHLGYVTREMLSVCYADTVEAVVAWLDGAPIRIANPQAVRRPPS*
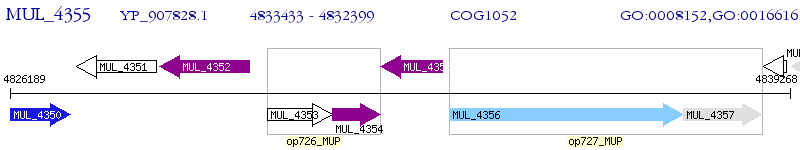
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4355 | serA4 | - | 100% (344) | D-3-phosphoglycerate dehydrogenase SerA4 |
| M. ulcerans Agy99 | MUL_1952 | serA1 | 2e-36 | 32.85% (274) | D-3-phosphoglycerate dehydrogenase |
| M. ulcerans Agy99 | MUL_1195 | serA3 | 3e-32 | 35.41% (257) | D-3-phosphoglycerate dehydrogenase SerA3 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3020c | serA1 | 2e-35 | 32.85% (274) | D-3-phosphoglycerate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4234 | - | 5e-38 | 33.21% (274) | D-3-phosphoglycerate dehydrogenase |
| M. tuberculosis H37Rv | Rv2996c | serA1 | 2e-35 | 32.85% (274) | D-3-phosphoglycerate dehydrogenase |
| M. leprae Br4923 | MLBr_01692 | serA | 7e-37 | 34.31% (274) | D-3-phosphoglycerate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3304c | - | 1e-35 | 32.12% (274) | D-3-phosphoglycerate dehydrogenase |
| M. marinum M | MMAR_5282 | serA4 | 0.0 | 98.55% (344) | D-3-phosphoglycerate dehydrogenase SerA4 |
| M. avium 104 | MAV_0349 | - | 1e-140 | 74.70% (328) | D-isomer specific 2-hydroxyacid dehydrogenase family protein |
| M. smegmatis MC2 155 | MSMEG_0604 | - | 4e-41 | 40.77% (260) | glyoxylate reductase |
| M. thermoresistible (build 8) | TH_2037 | serA | 3e-38 | 33.94% (274) | PROBABLE D-3-PHOSPHOGLYCERATE DEHYDROGENASE SERA1 (PGDH) |
| M. vanbaalenii PYR-1 | Mvan_2128 | - | 5e-38 | 33.21% (274) | D-3-phosphoglycerate dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4234|M.gilvum_PYR-GCK -----------MSLPVVLIADKLAQS-----TVEALGDQVEVRWVDG--P
Mvan_2128|M.vanbaalenii_PYR-1 -----------MSLPVVLIADKLAQS-----TVEALGDQVEVRWVDG--P
MAB_3304c|M.abscessus_ATCC_199 ----------------MLIADKLAPS-----TVEALGDGVEVRWVDG--P
Mb3020c|M.bovis_AF2122/97 -----------MSLPVVLIADKLAPS-----TVAALGDQVEVRWVDG--P
Rv2996c|M.tuberculosis_H37Rv -----------MSLPVVLIADKLAPS-----TVAALGDQVEVRWVDG--P
MLBr_01692|M.leprae_Br4923 -----------MDLPVVLIADKLAQS-----TVAALGDQVEVRWVDG--P
TH_2037|M.thermoresistible__bu -----------VSLPVVLIADKLAES-----TVAALGDQVEVRWVDG--P
MSMEG_0604|M.smegmatis_MC2_155 ---------MAVNPGTVRVLAHFAPSNTVLDFLAPHLDWLDVRFCGE--E
MUL_4355|M.ulcerans_Agy99 MSQPASHGFGQDHVPRVAILDDYARVATDLADWSPVRGRATLTVFDRHLG
MMAR_5282|M.marinum_M MSQPASHGFGQDHVPRVAILDDYARVATDLADWSPVRGRATLTVFDRHLG
MAV_0349|M.avium_104 -------------MPRLAILDDYARVALQVADWSRVQKRCDITVFDEHLS
: : . * : .
Mflv_4234|M.gilvum_PYR-GCK DREKLLAAVADADALLVRSATTVDAEVLAAAPKLKIVARAGVGLDNVDVD
Mvan_2128|M.vanbaalenii_PYR-1 DREKLLAAVADADALLVRSATTVDAEVLAAAPKLKIVARAGVGLDNVDVD
MAB_3304c|M.abscessus_ATCC_199 DRPALLAAVPEADALLVRSATTVDAEVLAAGTKLKIVARAGVGLDNVDVK
Mb3020c|M.bovis_AF2122/97 DRDKLLAAVPEADALLVRSATTVDAEVLAAAPKLKIVARAGVGLDNVDVD
Rv2996c|M.tuberculosis_H37Rv DRDKLLAAVPEADALLVRSATTVDAEVLAAAPKLKIVARAGVGLDNVDVD
MLBr_01692|M.leprae_Br4923 DRTKLLAAVPEADALLVRSATTVDAEVLAAAPKLKIVARAGVGLDNVDVD
TH_2037|M.thermoresistible__bu DREKLLAAVADADALLVRSATTVDAEVLAAAPKLKIVARAGVGLDNVDVE
MSMEG_0604|M.smegmatis_MC2_155 DDETFYRELGDADVLWH-VLRPITGDDLNRAPRLRLVHKLGAGVNTIDVE
MUL_4355|M.ulcerans_Agy99 QAEAAEALADFDAVCTMRERMAFPRSLLERLPRLKLITIVGMNLPNLDLA
MMAR_5282|M.marinum_M QAEAAEALADFDAVCTMRERMAFPRSLLERLPRLKLITIVGMNLPNLDLA
MAV_0349|M.avium_104 EQQAVDALRPFDVVCTMRERMAFPRSLIERLPHLRLLTIVGRQLPNLDMA
: . .. . : .:*::: * : .:*:
Mflv_4234|M.gilvum_PYR-GCK AATARGVLVVNAPTSN------IHSAAEHALALLLSTARQIPAADATLRE
Mvan_2128|M.vanbaalenii_PYR-1 AATARGVLVVNAPTSN------IHSAAEHALALLLAAARQIPAADATLRE
MAB_3304c|M.abscessus_ATCC_199 AATARGVLVVNAPTSN------IHSAAEHAVTLLLATARQIPAADASLKA
Mb3020c|M.bovis_AF2122/97 AATARGVLVVNAPTSN------IHSAAEHALALLLAASRQIPAADASLRE
Rv2996c|M.tuberculosis_H37Rv AATARGVLVVNAPTSN------IHSAAEHALALLLAASRQIPAADASLRE
MLBr_01692|M.leprae_Br4923 AATARGVLVVNAPTSN------IHSAAEHALALLLAASRQIAEADASLRA
TH_2037|M.thermoresistible__bu AATARGVLVVNAPTSN------IHSAAEHAIALLLAAARQIPAADATLRQ
MSMEG_0604|M.smegmatis_MC2_155 TATQLGILVANMPGAN------APSVAEGTVLLMLAALRRLPQLDRATRA
MUL_4355|M.ulcerans_Agy99 AATERGVLVAHSNFAHPRFRAARDATAEFAWGLLIATVHNLAEEHRQVRA
MMAR_5282|M.marinum_M AATERGVLVAHSNFAHPRFRAARDATAEFAWGLLIATVRNLAEEHRQLRA
MAV_0349|M.avium_104 AATEHGIAVARSEFAHPRFAALRDATPELAWGLMIATVRHLADEHRQMRA
:** *: *.. :: :..* : *:::: :.:. . :
Mflv_4234|M.gilvum_PYR-GCK HT-WKRSAFSG---TEIFGKTVGVVGLGRIGQLVAQRLAAFGAHITAYDP
Mvan_2128|M.vanbaalenii_PYR-1 HS-WKRSSFSG---TEIFGKTVGVVGLGRIGQLVAQRLAAFGAHITAYDP
MAB_3304c|M.abscessus_ATCC_199 HT-WKRSSFNG---TEIFGKTVGVVGLGRIGQLFAQRLAAFGTHIVAYDP
Mb3020c|M.bovis_AF2122/97 HT-WKRSSFSG---TEIFGKTVGVVGLGRIGQLVAQRIAAFGAYVVAYDP
Rv2996c|M.tuberculosis_H37Rv HT-WKRSSFSG---TEIFGKTVGVVGLGRIGQLVAQRIAAFGAYVVAYDP
MLBr_01692|M.leprae_Br4923 HI-WKRSSFSG---TEIFGKTVGVVGLGRIGQLVAARIAAFGAHVIAYDP
TH_2037|M.thermoresistible__bu RT-WKRSEFSG---VEIYDKTVGVVGLGRIGQLVAQRLAAFGTHLIAYDP
MSMEG_0604|M.smegmatis_MC2_155 GRGWPTDPTLGDTVRDIGGCTVGLVGYGNVAKRVERIVLAMGAEQVLHTS
MUL_4355|M.ulcerans_Agy99 GG-WQTTTGMT-----LSGKTLGLLGLGRVGKRMTEYAKAFGMDVIAWSQ
MMAR_5282|M.marinum_M GG-WQTTTGMT-----LSGKTLGLLGLGRVGKRMTEYAKAFGMDVIAWSQ
MAV_0349|M.avium_104 GG-WQRTAGMT-----LSGKTLGLLGLGRVGSRMAGFAAAFGMEVIAWSQ
* : . *:*::* *.:.. . *:*
Mflv_4234|M.gilvum_PYR-GCK YVSHARAAQLGIELLTLDELLSRADFISVHLPKTKETAGLIGTEALAKTK
Mvan_2128|M.vanbaalenii_PYR-1 YVSHARAAQLGIELLTLDELLGRADFISVHLPKTKETAGLIGKEALAKTK
MAB_3304c|M.abscessus_ATCC_199 YVSAARAAQLGIELLTLDELLGRADFISVHLPKTPETAGLIGKEALAKTK
Mb3020c|M.bovis_AF2122/97 YVSPARAAQLGIELLSLDDLLARADFISVHLPKTPETAGLIDKEALAKTK
Rv2996c|M.tuberculosis_H37Rv YVSPARAAQLGIELLSLDDLLARADFISVHLPKTPETAGLIDKEALAKTK
MLBr_01692|M.leprae_Br4923 YVAPARAAQLGIELMSFDDLLARADFISVHLPKTPETAGLIDKEALAKTK
TH_2037|M.thermoresistible__bu YVSPARAAQLGIELVDLDELLGRADIISVHLPKTPETAGLIGADALAKVK
MSMEG_0604|M.smegmatis_MC2_155 TRDTGHPRWR-----NLPDLLAASDIVSLHLPLTDTSRGLLGPEAIAAMK
MUL_4355|M.ulcerans_Agy99 NLTEEAAAAHGVRRVDKPVLFAQSDMVSIHVVLSERTRALVGAGELAAMK
MMAR_5282|M.marinum_M NLTEEAAAAHGVRRVDKPVLFAQSDVVSIHVVLSERTRALVGAGELAAMK
MAV_0349|M.avium_104 NLTDDAAAAVGARRVSKAALFESADVVSVHLVLSERTRGLVGEPELALMK
. *: :*.:*:*: : : .*:. :* *
Mflv_4234|M.gilvum_PYR-GCK PGVIIVNAARGGLIDEQALADAITSGHVRGAGLDVFSTEPC-TDSPLFEL
Mvan_2128|M.vanbaalenii_PYR-1 PGVIIVNAARGGLIDEAALADAINSGHVRGAGLDVFSTEPC-TDSPLFEL
MAB_3304c|M.abscessus_ATCC_199 PGVIIVNAARGGLIDEAALADAIRSGHVRGAGLDVFSTEPC-TDSPLFEL
Mb3020c|M.bovis_AF2122/97 PGVIIVNAARGGLVDEAALADAITGGHVRAAGLDVFATEPC-TDSPLFEL
Rv2996c|M.tuberculosis_H37Rv PGVIIVNAARGGLVDEAALADAITGGHVRAAGLDVFATEPC-TDSPLFEL
MLBr_01692|M.leprae_Br4923 PGVIIVNAARGGLVDEVALADAVRSGHVRAAGLDVFATEPC-TDSPLFEL
TH_2037|M.thermoresistible__bu PGVIIVNAARGGLIDEQALADAIISGRVRAAGIDVFAKEPC-TDSPLFDL
MSMEG_0604|M.smegmatis_MC2_155 PGAVLVNTARGPIVDEAALIEALRGGRLAAAGLDVFDTEPLPADHPLLGL
MUL_4355|M.ulcerans_Agy99 PGAYLINTSRGPIVDEGALIGALEAGRIAGAGLDVYDVEPLPVDHRLRSL
MMAR_5282|M.marinum_M PGAYLINTSRGPIVDEGALIGALEAGRIAGAGLDVYDVEPLPVDHRLRSL
MAV_0349|M.avium_104 PSAYLINTARGPIVDEPALIAALTAGRIAGAGLDVYDVEPLPGDHPLRSL
*.. ::*::** ::** ** *: .*:: .**:**: ** * * *
Mflv_4234|M.gilvum_PYR-GCK PQVVVTPHLGASTVEAQDRAGTDVAASVKLALAGEFVPDAVNVGGGAVGE
Mvan_2128|M.vanbaalenii_PYR-1 PQVVVTPHLGASTVEAQDRAGTDVAASVKLALAGEFVPDAVNVGGGAVGE
MAB_3304c|M.abscessus_ATCC_199 DQVVVTPHLGASTSEAQDRAGTDVAASVQLALAGEFVPDAVNVGGGVVSE
Mb3020c|M.bovis_AF2122/97 AQVVVTPHLGASTAEAQDRAGTDVAESVRLALAGEFVPDAVNVGGGVVNE
Rv2996c|M.tuberculosis_H37Rv AQVVVTPHLGASTAEAQDRAGTDVAESVRLALAGEFVPDAVNVGGGVVNE
MLBr_01692|M.leprae_Br4923 SQVVVTPHLGASTAEAQDRAGTDVAESVRLALAGEFVPDAVNVDGGVVNE
TH_2037|M.thermoresistible__bu PQVVVTPHLGASTVEAQDRAGTDVAASVKLALAGEFVPDAVNVGAGEVSE
MSMEG_0604|M.smegmatis_MC2_155 DNVVLTPHVTWYTADTMRRYLSIGVENCRRIRDGEPLAHLVNEAR-----
MUL_4355|M.ulcerans_Agy99 PNVTLSPHLGYVTREMLSVCYADTVEAVVAWLDGAPIRIANPQAVRRPPS
MMAR_5282|M.marinum_M PNVTLSPHLGYVTREMLSACYADTVEAVVAWLDGAPIRIANPQAVQRPPS
MAV_0349|M.avium_104 PNVTLSPHLGYVTREMLGAFYADTVESVLAWLDGTPIRIANPEVLQRD--
:*.::**: * : : . * :
Mflv_4234|M.gilvum_PYR-GCK EVAPWLDLVRKLGLLVGALSAELPTNLCVQVRGELASEEVEVLRLSALRG
Mvan_2128|M.vanbaalenii_PYR-1 EVAPWLDLVRKLGLLVGVLSSEPPVSLQVQVQGELASEEVEVLKLSALRG
MAB_3304c|M.abscessus_ATCC_199 EVAPWLEVVRKLGVLIGAVSEQLPTSLSVDVRGELASEDVAVLKLSALRG
Mb3020c|M.bovis_AF2122/97 EVAPWLDLVRKLGVLAGVLSDELPVSLSVQVRGELAAEEVEVLRLSALRG
Rv2996c|M.tuberculosis_H37Rv EVAPWLDLVRKLGVLAGVLSDELPVSLSVQVRGELAAEEVEVLRLSALRG
MLBr_01692|M.leprae_Br4923 EVAPWLDLVCKLGVLVAALSDELPASLSVHVRGELASEDVEILRLSALRG
TH_2037|M.thermoresistible__bu EVAPWVDLARKLGLLVGTLSGEPTATLQVRVAGELASEDVEVLRLSALRG
MSMEG_0604|M.smegmatis_MC2_155 --------------------------------------------------
MUL_4355|M.ulcerans_Agy99 --------------------------------------------------
MMAR_5282|M.marinum_M --------------------------------------------------
MAV_0349|M.avium_104 --------------------------------------------------
Mflv_4234|M.gilvum_PYR-GCK LFSAVIEDQVTFVNAPALATERGVEASIDTESESPNHRSVVDVRAVAADG
Mvan_2128|M.vanbaalenii_PYR-1 LFSAVIEHPVTFVNAPALASERGVEASITTATESANHRSVVDVRAVAADG
MAB_3304c|M.abscessus_ATCC_199 LFSSVIEDQVTFVNAPSIAEERGVSAEITTATESPNHRSVVDVRAVYADG
Mb3020c|M.bovis_AF2122/97 LFSAVIEDAVTFVNAPALAAERGVTAEICKASESPNHRSVVDVRAVGADG
Rv2996c|M.tuberculosis_H37Rv LFSAVIEDAVTFVNAPALAAERGVTAEICKASESPNHRSVVDVRAVGADG
MLBr_01692|M.leprae_Br4923 LFSTVIEDAVTFVNAPALAAERGVSAEITTGSESPNHRSVVDVRAVASDG
TH_2037|M.thermoresistible__bu LFSTLTDQQVTYVNAPTLAAERGMQAELEKVSESPNYRSVVDVRAVSTDG
MSMEG_0604|M.smegmatis_MC2_155 --------------------------------------------------
MUL_4355|M.ulcerans_Agy99 --------------------------------------------------
MMAR_5282|M.marinum_M --------------------------------------------------
MAV_0349|M.avium_104 --------------------------------------------------
Mflv_4234|M.gilvum_PYR-GCK STVNVSGTLTGPQLVEKIVQINGRNLDLRAEGVNVIINYHDQPGALGKIG
Mvan_2128|M.vanbaalenii_PYR-1 STVNVAGTLTGPQLVEKIVQINGRNLELRAEGVNLIINYDDQPGALGKIG
MAB_3304c|M.abscessus_ATCC_199 SVINVAGTLSGPQLVQKIVQINGRNFDLRAEGVNLVINYADVPGALGKIG
Mb3020c|M.bovis_AF2122/97 SVVTVSGTLYGPQLSQKIVQINGRHFDLRAQGINLIIHYVDRPGALGKIG
Rv2996c|M.tuberculosis_H37Rv SVVTVSGTLYGPQLSQKIVQINGRHFDLRAQGINLIIHYVDRPGALGKIG
MLBr_01692|M.leprae_Br4923 SVVNIAGTLSGPQLVQKIVQVNGRNFDLRAQGMNLVIRYVDQPGALGKIG
TH_2037|M.thermoresistible__bu TVTNVAGTLSGPQQVEKIVQINGRNFDLRAEGVNLVMHYVDRPGALGKIG
MSMEG_0604|M.smegmatis_MC2_155 --------------------------------------------------
MUL_4355|M.ulcerans_Agy99 --------------------------------------------------
MMAR_5282|M.marinum_M --------------------------------------------------
MAV_0349|M.avium_104 --------------------------------------------------
Mflv_4234|M.gilvum_PYR-GCK TLLGGANVNILAAQLSQDADGEGATIMLRVDREVPADVLAGIGRDVNALT
Mvan_2128|M.vanbaalenii_PYR-1 TLLGGAAVNILAAQLSQDADGIGATVMLRLDREVPGEVLAAIGRDVNAVT
MAB_3304c|M.abscessus_ATCC_199 TVLGGAEVNIQAAQLSQDASGAAATIILRIDRTAPDAVLDEIRAAVGATT
Mb3020c|M.bovis_AF2122/97 TLLGTAGVNIQAAQLSEDAEGPGATILLRLDQDVPDDVRTAIAAAVDAYK
Rv2996c|M.tuberculosis_H37Rv TLLGTAGVNIQAAQLSEDAEGPGATILLRLDQDVPDDVRTAIAAAVDAYK
MLBr_01692|M.leprae_Br4923 TLLGAAGVNIQAAQLSEDTEGPGATILLRLDQDVPGDVRSAIVAAVSANK
TH_2037|M.thermoresistible__bu TLLGAADVNILAAQLSQDISGDGATLMLRVNRDVPADVRDAIGAAVGAST
MSMEG_0604|M.smegmatis_MC2_155 --------------------------------------------------
MUL_4355|M.ulcerans_Agy99 --------------------------------------------------
MMAR_5282|M.marinum_M --------------------------------------------------
MAV_0349|M.avium_104 --------------------------------------------------
Mflv_4234|M.gilvum_PYR-GCK LEVVDLT
Mvan_2128|M.vanbaalenii_PYR-1 LEVVDLT
MAB_3304c|M.abscessus_ATCC_199 LELVDLS
Mb3020c|M.bovis_AF2122/97 LEVVDLS
Rv2996c|M.tuberculosis_H37Rv LEVVDLS
MLBr_01692|M.leprae_Br4923 LEVVNLS
TH_2037|M.thermoresistible__bu LEVVDLS
MSMEG_0604|M.smegmatis_MC2_155 -------
MUL_4355|M.ulcerans_Agy99 -------
MMAR_5282|M.marinum_M -------
MAV_0349|M.avium_104 -------