For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VPRLAILDDYARVALQVADWSRVQKRCDITVFDEHLSEQQAVDALRPFDVVCTMRERMAFPRSLIERLPH LRLLTIVGRQLPNLDMAAATEHGIAVARSEFAHPRFAALRDATPELAWGLMIATVRHLADEHRQMRAGGW QRTAGMTLSGKTLGLLGLGRVGSRMAGFAAAFGMEVIAWSQNLTDDAAAAVGARRVSKAALFESADVVSV HLVLSERTRGLVGEPELALMKPSAYLINTARGPIVDEPALIAALTAGRIAGAGLDVYDVEPLPGDHPLRS LPNVTLSPHLGYVTREMLGAFYADTVESVLAWLDGTPIRIANPEVLQRD
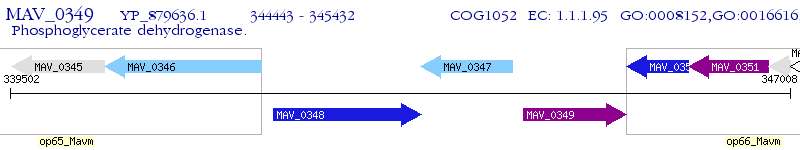
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0349 | - | - | 100% (329) | D-isomer specific 2-hydroxyacid dehydrogenase family protein |
| M. avium 104 | MAV_3847 | serA | 4e-36 | 34.85% (264) | D-3-phosphoglycerate dehydrogenase |
| M. avium 104 | MAV_4438 | - | 3e-32 | 36.19% (257) | 2-hydroxyacid dehydrogenase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3020c | serA1 | 6e-36 | 35.23% (264) | D-3-phosphoglycerate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4234 | - | 8e-38 | 35.23% (264) | D-3-phosphoglycerate dehydrogenase |
| M. tuberculosis H37Rv | Rv2996c | serA1 | 6e-36 | 35.23% (264) | D-3-phosphoglycerate dehydrogenase |
| M. leprae Br4923 | MLBr_01692 | serA | 3e-37 | 35.98% (264) | D-3-phosphoglycerate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3304c | - | 9e-36 | 36.03% (247) | D-3-phosphoglycerate dehydrogenase |
| M. marinum M | MMAR_5282 | serA4 | 1e-142 | 75.91% (328) | D-3-phosphoglycerate dehydrogenase SerA4 |
| M. smegmatis MC2 155 | MSMEG_0604 | - | 3e-40 | 40.30% (263) | glyoxylate reductase |
| M. thermoresistible (build 8) | TH_3809 | - | 2e-40 | 37.36% (273) | PUTATIVE putative D-3-phosphoglycerate dehydrogenase |
| M. ulcerans Agy99 | MUL_4355 | serA4 | 1e-140 | 74.70% (328) | D-3-phosphoglycerate dehydrogenase SerA4 |
| M. vanbaalenii PYR-1 | Mvan_2128 | - | 8e-37 | 34.47% (264) | D-3-phosphoglycerate dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4234|M.gilvum_PYR-GCK ------------MSLPVVLIADKLAQS-----TVEALGDQVEVRWVDGPD
Mvan_2128|M.vanbaalenii_PYR-1 ------------MSLPVVLIADKLAQS-----TVEALGDQVEVRWVDGPD
MAB_3304c|M.abscessus_ATCC_199 -----------------MLIADKLAPS-----TVEALGDGVEVRWVDGPD
Mb3020c|M.bovis_AF2122/97 ------------MSLPVVLIADKLAPS-----TVAALGDQVEVRWVDGPD
Rv2996c|M.tuberculosis_H37Rv ------------MSLPVVLIADKLAPS-----TVAALGDQVEVRWVDGPD
MLBr_01692|M.leprae_Br4923 ------------MDLPVVLIADKLAQS-----TVAALGDQVEVRWVDGPD
MSMEG_0604|M.smegmatis_MC2_155 ----------MAVNPGTVRVLAHFAPSNTVLDFLAPHLDWLDVRFCGEED
TH_3809|M.thermoresistible__bu ---------VSGISRPRLVFERWTDPAAG---ELLGAADIELVRLDLTAD
MMAR_5282|M.marinum_M MSQPASHGFGQDHVPRVAILDDYARVATDLADWSPVRGRATLTVFDRHLG
MUL_4355|M.ulcerans_Agy99 MSQPASHGFGQDHVPRVAILDDYARVATDLADWSPVRGRATLTVFDRHLG
MAV_0349|M.avium_104 -------------MPRLAILDDYARVALQVADWSRVQKRCDITVFDEHLS
. : . .
Mflv_4234|M.gilvum_PYR-GCK REKLLAAVADADALLVRSATTVD-----------AEVLAAAPKLKIVARA
Mvan_2128|M.vanbaalenii_PYR-1 REKLLAAVADADALLVRSATTVD-----------AEVLAAAPKLKIVARA
MAB_3304c|M.abscessus_ATCC_199 RPALLAAVPEADALLVRSATTVD-----------AEVLAAGTKLKIVARA
Mb3020c|M.bovis_AF2122/97 RDKLLAAVPEADALLVRSATTVD-----------AEVLAAAPKLKIVARA
Rv2996c|M.tuberculosis_H37Rv RDKLLAAVPEADALLVRSATTVD-----------AEVLAAAPKLKIVARA
MLBr_01692|M.leprae_Br4923 RTKLLAAVPEADALLVRSATTVD-----------AEVLAAAPKLKIVARA
MSMEG_0604|M.smegmatis_MC2_155 DETFYRELGDADVLWH-VLRPIT-----------GDDLNRAPRLRLVHKL
TH_3809|M.thermoresistible__bu PADGWAALESAHGYQVATRTDVASMADGGQWLADRNLVRRCGLLLAVCSA
MMAR_5282|M.marinum_M QAEAAEALADFDAVCTMRERMAFP----------RSLLERLPRLKLITIV
MUL_4355|M.ulcerans_Agy99 QAEAAEALADFDAVCTMRERMAFP----------RSLLERLPRLKLITIV
MAV_0349|M.avium_104 EQQAVDALRPFDVVCTMRERMAFP----------RSLIERLPHLRLLTIV
: . . : * :
Mflv_4234|M.gilvum_PYR-GCK GVGLDNVDVDAATARGVLVVNAPTSN------IHSAAEHALALLLSTARQ
Mvan_2128|M.vanbaalenii_PYR-1 GVGLDNVDVDAATARGVLVVNAPTSN------IHSAAEHALALLLAAARQ
MAB_3304c|M.abscessus_ATCC_199 GVGLDNVDVKAATARGVLVVNAPTSN------IHSAAEHAVTLLLATARQ
Mb3020c|M.bovis_AF2122/97 GVGLDNVDVDAATARGVLVVNAPTSN------IHSAAEHALALLLAASRQ
Rv2996c|M.tuberculosis_H37Rv GVGLDNVDVDAATARGVLVVNAPTSN------IHSAAEHALALLLAASRQ
MLBr_01692|M.leprae_Br4923 GVGLDNVDVDAATARGVLVVNAPTSN------IHSAAEHALALLLAASRQ
MSMEG_0604|M.smegmatis_MC2_155 GAGVNTIDVETATQLGILVANMPGAN------APSVAEGTVLLMLAALRR
TH_3809|M.thermoresistible__bu GAGYDVIDVDACTEAGIAVCNNSGPG------AEAVAEHALGLMLDLAKR
MMAR_5282|M.marinum_M GMNLPNLDLAAATERGVLVAHSNFAHPRFRAARDATAEFAWGLLIATVRN
MUL_4355|M.ulcerans_Agy99 GMNLPNLDLAAATERGVLVAHSNFAHPRFRAARDATAEFAWGLLIATVHN
MAV_0349|M.avium_104 GRQLPNLDMAAATEHGIAVARSEFAHPRFAALRDATPELAWGLMIATVRH
* :*: :.* *: * . . :..* : *:: :.
Mflv_4234|M.gilvum_PYR-GCK IPAADATLREHT-WKRSAFSG---TEIFGKTVGVVGLGRIGQLVAQRLAA
Mvan_2128|M.vanbaalenii_PYR-1 IPAADATLREHS-WKRSSFSG---TEIFGKTVGVVGLGRIGQLVAQRLAA
MAB_3304c|M.abscessus_ATCC_199 IPAADASLKAHT-WKRSSFNG---TEIFGKTVGVVGLGRIGQLFAQRLAA
Mb3020c|M.bovis_AF2122/97 IPAADASLREHT-WKRSSFSG---TEIFGKTVGVVGLGRIGQLVAQRIAA
Rv2996c|M.tuberculosis_H37Rv IPAADASLREHT-WKRSSFSG---TEIFGKTVGVVGLGRIGQLVAQRIAA
MLBr_01692|M.leprae_Br4923 IAEADASLRAHI-WKRSSFSG---TEIFGKTVGVVGLGRIGQLVAARIAA
MSMEG_0604|M.smegmatis_MC2_155 LPQLDRATRAGRGWPTDPTLGDTVRDIGGCTVGLVGYGNVAKRVERIVLA
TH_3809|M.thermoresistible__bu ITVADRMLRRGDLGDRLVLRG---SQLQHKTMGIVGLGAIGRRLVQLVAP
MMAR_5282|M.marinum_M LAEEHRQLRAGG---WQTTTG---MTLSGKTLGLLGLGRVGKRMTEYAKA
MUL_4355|M.ulcerans_Agy99 LAEEHRQVRAGG---WQTTTG---MTLSGKTLGLLGLGRVGKRMTEYAKA
MAV_0349|M.avium_104 LADEHRQMRAGG---WQRTAG---MTLSGKTLGLLGLGRVGSRMAGFAAA
:. . : * : *:*::* * :. . .
Mflv_4234|M.gilvum_PYR-GCK FGAHITAYDPYVSHARAAQLGIELLTLDELLSRADFISVHLPKTKETAGL
Mvan_2128|M.vanbaalenii_PYR-1 FGAHITAYDPYVSHARAAQLGIELLTLDELLGRADFISVHLPKTKETAGL
MAB_3304c|M.abscessus_ATCC_199 FGTHIVAYDPYVSAARAAQLGIELLTLDELLGRADFISVHLPKTPETAGL
Mb3020c|M.bovis_AF2122/97 FGAYVVAYDPYVSPARAAQLGIELLSLDDLLARADFISVHLPKTPETAGL
Rv2996c|M.tuberculosis_H37Rv FGAYVVAYDPYVSPARAAQLGIELLSLDDLLARADFISVHLPKTPETAGL
MLBr_01692|M.leprae_Br4923 FGAHVIAYDPYVAPARAAQLGIELMSFDDLLARADFISVHLPKTPETAGL
MSMEG_0604|M.smegmatis_MC2_155 MGAEQVLHTSTRDTGHPRWR-----NLPDLLAASDIVSLHLPLTDTSRGL
TH_3809|M.thermoresistible__bu FGMKVLAFDPYVDEKTAEALGARSVSLEELLGCSDFVQLTCPLTAETEGL
MMAR_5282|M.marinum_M FGMDVIAWSQNLTEEAAAAHGVRRVDKPVLFAQSDVVSIHVVLSERTRAL
MUL_4355|M.ulcerans_Agy99 FGMDVIAWSQNLTEEAAAAHGVRRVDKPVLFAQSDMVSIHVVLSERTRAL
MAV_0349|M.avium_104 FGMEVIAWSQNLTDDAAAAVGARRVSKAALFESADVVSVHLVLSERTRGL
:* . *: :*.:.: : : .*
Mflv_4234|M.gilvum_PYR-GCK IGTEALAKTKPGVIIVNAARGGLIDEQALADAITSGHVRGAGLDVFSTEP
Mvan_2128|M.vanbaalenii_PYR-1 IGKEALAKTKPGVIIVNAARGGLIDEAALADAINSGHVRGAGLDVFSTEP
MAB_3304c|M.abscessus_ATCC_199 IGKEALAKTKPGVIIVNAARGGLIDEAALADAIRSGHVRGAGLDVFSTEP
Mb3020c|M.bovis_AF2122/97 IDKEALAKTKPGVIIVNAARGGLVDEAALADAITGGHVRAAGLDVFATEP
Rv2996c|M.tuberculosis_H37Rv IDKEALAKTKPGVIIVNAARGGLVDEAALADAITGGHVRAAGLDVFATEP
MLBr_01692|M.leprae_Br4923 IDKEALAKTKPGVIIVNAARGGLVDEVALADAVRSGHVRAAGLDVFATEP
MSMEG_0604|M.smegmatis_MC2_155 LGPEAIAAMKPGAVLVNTARGPIVDEAALIEALRGGRLAAAGLDVFDTEP
TH_3809|M.thermoresistible__bu IGRAQFAAMKPTAFFVTTARGRVHDEAALYEALTCGEIAGAGLDVFHDEP
MMAR_5282|M.marinum_M VGAGELAAMKPGAYLINTSRGPIVDEGALIGALEAGRIAGAGLDVYDVEP
MUL_4355|M.ulcerans_Agy99 VGAGELAAMKPGAYLINTSRGPIVDEGALIGALEAGRIAGAGLDVYDVEP
MAV_0349|M.avium_104 VGEPELALMKPSAYLINTARGPIVDEPALIAALTAGRIAGAGLDVYDVEP
:. :* ** . ::.::** : ** ** *: *.: .*****: **
Mflv_4234|M.gilvum_PYR-GCK C-TDSPLFELPQVVVTPHLGASTVEAQDRAGTDVAASVKLALAGEFVPDA
Mvan_2128|M.vanbaalenii_PYR-1 C-TDSPLFELPQVVVTPHLGASTVEAQDRAGTDVAASVKLALAGEFVPDA
MAB_3304c|M.abscessus_ATCC_199 C-TDSPLFELDQVVVTPHLGASTSEAQDRAGTDVAASVQLALAGEFVPDA
Mb3020c|M.bovis_AF2122/97 C-TDSPLFELAQVVVTPHLGASTAEAQDRAGTDVAESVRLALAGEFVPDA
Rv2996c|M.tuberculosis_H37Rv C-TDSPLFELAQVVVTPHLGASTAEAQDRAGTDVAESVRLALAGEFVPDA
MLBr_01692|M.leprae_Br4923 C-TDSPLFELSQVVVTPHLGASTAEAQDRAGTDVAESVRLALAGEFVPDA
MSMEG_0604|M.smegmatis_MC2_155 LPADHPLLGLDNVVLTPHVTWYTADTMRRYLSIGVENCRRIRDGEPLAHL
TH_3809|M.thermoresistible__bu PRADHPLLTLDNVVATPHIAGITAEAARDIAVATADQWQTIFAGGVPPRL
MMAR_5282|M.marinum_M LPVDHRLRSLPNVTLSPHLGYVTREMLSACYADTVEAVVAWLDGAPIRIA
MUL_4355|M.ulcerans_Agy99 LPVDHRLRSLPNVTLSPHLGYVTREMLSVCYADTVEAVVAWLDGAPIRIA
MAV_0349|M.avium_104 LPGDHPLRSLPNVTLSPHLGYVTREMLGAFYADTVESVLAWLDGTPIRIA
* * * :*. :**: * : . *
Mflv_4234|M.gilvum_PYR-GCK VNVGGGAVGEEVAPWLDLVRKLGLLVGALSAELPTNLCVQVRGELASEEV
Mvan_2128|M.vanbaalenii_PYR-1 VNVGGGAVGEEVAPWLDLVRKLGLLVGVLSSEPPVSLQVQVQGELASEEV
MAB_3304c|M.abscessus_ATCC_199 VNVGGGVVSEEVAPWLEVVRKLGVLIGAVSEQLPTSLSVDVRGELASEDV
Mb3020c|M.bovis_AF2122/97 VNVGGGVVNEEVAPWLDLVRKLGVLAGVLSDELPVSLSVQVRGELAAEEV
Rv2996c|M.tuberculosis_H37Rv VNVGGGVVNEEVAPWLDLVRKLGVLAGVLSDELPVSLSVQVRGELAAEEV
MLBr_01692|M.leprae_Br4923 VNVDGGVVNEEVAPWLDLVCKLGVLVAALSDELPASLSVHVRGELASEDV
MSMEG_0604|M.smegmatis_MC2_155 VNEAR---------------------------------------------
TH_3809|M.thermoresistible__bu LNP---------EVWPRYSARFAEIFGFAPEPVSRTREREAVGR------
MMAR_5282|M.marinum_M NPQAVQRPPS----------------------------------------
MUL_4355|M.ulcerans_Agy99 NPQAVRRPPS----------------------------------------
MAV_0349|M.avium_104 NPEVLQRD------------------------------------------
Mflv_4234|M.gilvum_PYR-GCK EVLRLSALRGLFSAVIEDQVTFVNAPALATERGVEASIDTESESPNHRSV
Mvan_2128|M.vanbaalenii_PYR-1 EVLKLSALRGLFSAVIEHPVTFVNAPALASERGVEASITTATESANHRSV
MAB_3304c|M.abscessus_ATCC_199 AVLKLSALRGLFSSVIEDQVTFVNAPSIAEERGVSAEITTATESPNHRSV
Mb3020c|M.bovis_AF2122/97 EVLRLSALRGLFSAVIEDAVTFVNAPALAAERGVTAEICKASESPNHRSV
Rv2996c|M.tuberculosis_H37Rv EVLRLSALRGLFSAVIEDAVTFVNAPALAAERGVTAEICKASESPNHRSV
MLBr_01692|M.leprae_Br4923 EILRLSALRGLFSTVIEDAVTFVNAPALAAERGVSAEITTGSESPNHRSV
MSMEG_0604|M.smegmatis_MC2_155 --------------------------------------------------
TH_3809|M.thermoresistible__bu --------------------------------------------------
MMAR_5282|M.marinum_M --------------------------------------------------
MUL_4355|M.ulcerans_Agy99 --------------------------------------------------
MAV_0349|M.avium_104 --------------------------------------------------
Mflv_4234|M.gilvum_PYR-GCK VDVRAVAADGSTVNVSGTLTGPQLVEKIVQINGRNLDLRAEGVNVIINYH
Mvan_2128|M.vanbaalenii_PYR-1 VDVRAVAADGSTVNVAGTLTGPQLVEKIVQINGRNLELRAEGVNLIINYD
MAB_3304c|M.abscessus_ATCC_199 VDVRAVYADGSVINVAGTLSGPQLVQKIVQINGRNFDLRAEGVNLVINYA
Mb3020c|M.bovis_AF2122/97 VDVRAVGADGSVVTVSGTLYGPQLSQKIVQINGRHFDLRAQGINLIIHYV
Rv2996c|M.tuberculosis_H37Rv VDVRAVGADGSVVTVSGTLYGPQLSQKIVQINGRHFDLRAQGINLIIHYV
MLBr_01692|M.leprae_Br4923 VDVRAVASDGSVVNIAGTLSGPQLVQKIVQVNGRNFDLRAQGMNLVIRYV
MSMEG_0604|M.smegmatis_MC2_155 --------------------------------------------------
TH_3809|M.thermoresistible__bu --------------------------------------------------
MMAR_5282|M.marinum_M --------------------------------------------------
MUL_4355|M.ulcerans_Agy99 --------------------------------------------------
MAV_0349|M.avium_104 --------------------------------------------------
Mflv_4234|M.gilvum_PYR-GCK DQPGALGKIGTLLGGANVNILAAQLSQDADGEGATIMLRVDREVPADVLA
Mvan_2128|M.vanbaalenii_PYR-1 DQPGALGKIGTLLGGAAVNILAAQLSQDADGIGATVMLRLDREVPGEVLA
MAB_3304c|M.abscessus_ATCC_199 DVPGALGKIGTVLGGAEVNIQAAQLSQDASGAAATIILRIDRTAPDAVLD
Mb3020c|M.bovis_AF2122/97 DRPGALGKIGTLLGTAGVNIQAAQLSEDAEGPGATILLRLDQDVPDDVRT
Rv2996c|M.tuberculosis_H37Rv DRPGALGKIGTLLGTAGVNIQAAQLSEDAEGPGATILLRLDQDVPDDVRT
MLBr_01692|M.leprae_Br4923 DQPGALGKIGTLLGAAGVNIQAAQLSEDTEGPGATILLRLDQDVPGDVRS
MSMEG_0604|M.smegmatis_MC2_155 --------------------------------------------------
TH_3809|M.thermoresistible__bu --------------------------------------------------
MMAR_5282|M.marinum_M --------------------------------------------------
MUL_4355|M.ulcerans_Agy99 --------------------------------------------------
MAV_0349|M.avium_104 --------------------------------------------------
Mflv_4234|M.gilvum_PYR-GCK GIGRDVNALTLEVVDLT
Mvan_2128|M.vanbaalenii_PYR-1 AIGRDVNAVTLEVVDLT
MAB_3304c|M.abscessus_ATCC_199 EIRAAVGATTLELVDLS
Mb3020c|M.bovis_AF2122/97 AIAAAVDAYKLEVVDLS
Rv2996c|M.tuberculosis_H37Rv AIAAAVDAYKLEVVDLS
MLBr_01692|M.leprae_Br4923 AIVAAVSANKLEVVNLS
MSMEG_0604|M.smegmatis_MC2_155 -----------------
TH_3809|M.thermoresistible__bu -----------------
MMAR_5282|M.marinum_M -----------------
MUL_4355|M.ulcerans_Agy99 -----------------
MAV_0349|M.avium_104 -----------------