For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LVPTLDRVPTPNQSGAAGGRAVRDRRPAVAVLCAQESDRPPGLEALDVEFRYCTADTLAEAVRGARAMLL WDFFSKAVRDVWSAAGSVEWIHVTAAGVDTLLFDELRDSDVVVTNAHGVFDRPIAEYVLGAVIAHAKDTR RSLDLQRQHRWQHRETRRVTGSRALVVGTGGIGRETARLLRAAGLEVRGAGRTARDDDPDFGRVVASSDL SAEVGWCDHLILAVPLTDATRGLVDAAVLGAMKPDAHLVNIARGPVVDEAALIAALTEHRIGGATLDAFD VEPLPADHPLWDVPNVTITAHMSGDVVGWRDTLAEQYAENVRRWLAGEPLHNVVDKKLGYISQAGRGVR
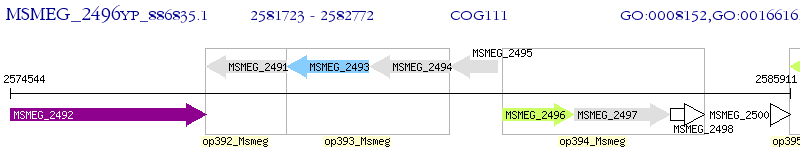
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2496 | - | - | 100% (349) | NAD-binding protein |
| M. smegmatis MC2 155 | MSMEG_3422 | - | 2e-36 | 38.34% (253) | D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding |
| M. smegmatis MC2 155 | MSMEG_0604 | - | 4e-31 | 33.65% (312) | glyoxylate reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3020c | serA1 | 1e-21 | 29.29% (297) | D-3-phosphoglycerate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4768 | - | 7e-28 | 36.04% (222) | D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding |
| M. tuberculosis H37Rv | Rv2996c | serA1 | 1e-21 | 29.29% (297) | D-3-phosphoglycerate dehydrogenase |
| M. leprae Br4923 | MLBr_01692 | serA | 2e-20 | 28.73% (268) | D-3-phosphoglycerate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4414 | - | 1e-71 | 45.86% (314) | putative NAD-binding protein (D-isomer specific 2-hydroxyacid |
| M. marinum M | MMAR_5282 | serA4 | 3e-27 | 33.33% (249) | D-3-phosphoglycerate dehydrogenase SerA4 |
| M. avium 104 | MAV_0349 | - | 7e-29 | 40.00% (205) | D-isomer specific 2-hydroxyacid dehydrogenase family protein |
| M. thermoresistible (build 8) | TH_3383 | - | 1e-26 | 31.69% (325) | PUTATIVE D-isomer specific 2-hydroxyacid dehydrogenase, |
| M. ulcerans Agy99 | MUL_4355 | serA4 | 4e-27 | 33.33% (249) | D-3-phosphoglycerate dehydrogenase SerA4 |
| M. vanbaalenii PYR-1 | Mvan_0404 | - | 5e-29 | 33.44% (302) | D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding |
CLUSTAL 2.0.9 multiple sequence alignment
TH_3383|M.thermoresistible__bu VDRPAVALSANRRRHCPGMLRGDMSGVNSRSALRVLAHFRPG---PKVLE
Mvan_0404|M.vanbaalenii_PYR-1 --------------------------------MRVLAHFVPGGPDSKVIE
Mb3020c|M.bovis_AF2122/97 ----------------------------------MSLPVVLIADKLAPST
Rv2996c|M.tuberculosis_H37Rv ----------------------------------MSLPVVLIADKLAPST
MLBr_01692|M.leprae_Br4923 ----------------------------------MDLPVVLIADKLAQST
Mflv_4768|M.gilvum_PYR-GCK ----------------------------------MQVELPRHRKRIEDLG
MMAR_5282|M.marinum_M ----------------MSQPASHGFGQDHVPRVAILDDYARVATDLADWS
MUL_4355|M.ulcerans_Agy99 ----------------MSQPASHGFGQDHVPRVAILDDYARVATDLADWS
MAV_0349|M.avium_104 -----------------------------MPRLAILDDYARVALQVADWS
MSMEG_2496|M.smegmatis_MC2_155 ----------MVPTLDRVPTPNQSGAAGGRAVRDRRPAVAVLCAQESDRP
MAB_4414|M.abscessus_ATCC_1997 --------------------------MNEQLVR--RPVIAVQHSPDS-VP
TH_3383|M.thermoresistible__bu FLEPESGWLDVRFCDEDDDETFHRELPHAEVIWHVLRPLSGEDLTRATRC
Mvan_0404|M.vanbaalenii_PYR-1 FLAPCRDWLDVRFCAEDDDDTFYRELADAEVIWHVLRPLSGEDLARAPRL
Mb3020c|M.bovis_AF2122/97 VAALGDQVEVRWVDGPDRDKLLAAVPEADALLVRSATTVDAEVLAAAPKL
Rv2996c|M.tuberculosis_H37Rv VAALGDQVEVRWVDGPDRDKLLAAVPEADALLVRSATTVDAEVLAAAPKL
MLBr_01692|M.leprae_Br4923 VAALGDQVEVRWVDGPDRTKLLAAVPEADALLVRSATTVDAEVLAAAPKL
Mflv_4768|M.gilvum_PYR-GCK FEVLAPELGGRQQFSSSE---LLEYSSRLIGIIAGDDELDADFFAGAGKL
MMAR_5282|M.marinum_M PVRGRATLTVFDRHLGQAEAAEALADFDAVCTMRERMAFPRSLLERLPRL
MUL_4355|M.ulcerans_Agy99 PVRGRATLTVFDRHLGQAEAAEALADFDAVCTMRERMAFPRSLLERLPRL
MAV_0349|M.avium_104 RVQKRCDITVFDEHLSEQQAVDALRPFDVVCTMRERMAFPRSLIERLPHL
MSMEG_2496|M.smegmatis_MC2_155 PGLEALD--VEFRYCTADTLAEAVRGARAMLLWDFFSKAVRDVWSAAGSV
MAB_4414|M.abscessus_ATCC_1997 ERLAPVTAAAEVRLVESDRLATALPGAEVLFVYDFRSSALRETWSAADSL
.
TH_3383|M.thermoresistible__bu RLVHKLGTGVNTIDVDTATERGIAVANMPGAN------AASVAEGTVMLM
Mvan_0404|M.vanbaalenii_PYR-1 RLVHKLGAGVNTIDVDAATARGVAVANMPGAN------AESVAEGTLLLM
Mb3020c|M.bovis_AF2122/97 KIVARAGVGLDNVDVDAATARGVLVVNAPTSN------IHSAAEHALALL
Rv2996c|M.tuberculosis_H37Rv KIVARAGVGLDNVDVDAATARGVLVVNAPTSN------IHSAAEHALALL
MLBr_01692|M.leprae_Br4923 KIVARAGVGLDNVDVDAATARGVLVVNAPTSN------IHSAAEHALALL
Mflv_4768|M.gilvum_PYR-GCK KTVIRWGIGMDSVDHEAARRHGVTVRNTPGVF------GYEVADSAFGYI
MMAR_5282|M.marinum_M KLITIVGMNLPNLDLAAATERGVLVAHSNFAHPRFRAARDATAEFAWGLL
MUL_4355|M.ulcerans_Agy99 KLITIVGMNLPNLDLAAATERGVLVAHSNFAHPRFRAARDATAEFAWGLL
MAV_0349|M.avium_104 RLLTIVGRQLPNLDMAAATEHGIAVARSEFAHPRFAALRDATPELAWGLM
MSMEG_2496|M.smegmatis_MC2_155 EWIHVTAAGVDTLLFDELRDSDVVVTNAHGVF------DRPIAEYVLGAV
MAB_4414|M.abscessus_ATCC_1997 RWVQIASTGVDPVLFDEFVASDVVLTNSRGIF------ERPIAEYVLAQI
. : . : : .: : . .: . :
TH_3383|M.thermoresistible__bu LAALRRLVELDRATRAGTGWPTDPSLGETVRDIGGSTVGLIGYGNVARRV
Mvan_0404|M.vanbaalenii_PYR-1 LAALRRLPVLDRATREGRGWPSDPTLGETVRDIGGCTVGLVGYGNIAKQV
Mb3020c|M.bovis_AF2122/97 LAASRQIPAADASLREH-TWKRSSFSGTEIF---GKTVGVVGLGRIGQLV
Rv2996c|M.tuberculosis_H37Rv LAASRQIPAADASLREH-TWKRSSFSGTEIF---GKTVGVVGLGRIGQLV
MLBr_01692|M.leprae_Br4923 LAASRQIAEADASLRAH-IWKRSSFSGTEIF---GKTVGVVGLGRIGQLV
Mflv_4768|M.gilvum_PYR-GCK LNLARGYMAVDAAVRRG-EWP--KVEGITLD---GSRLGIVGFGAIGREI
MMAR_5282|M.marinum_M IATVRNLAEEHRQLRAG-GWQT--TTGMTLS---GKTLGLLGLGRVGKRM
MUL_4355|M.ulcerans_Agy99 IATVHNLAEEHRQVRAG-GWQT--TTGMTLS---GKTLGLLGLGRVGKRM
MAV_0349|M.avium_104 IATVRHLADEHRQMRAG-GWQR--TAGMTLS---GKTLGLLGLGRVGSRM
MSMEG_2496|M.smegmatis_MC2_155 IAHAKDTRRSLDLQRQH-RWQHRETRRVTGS-----RALVVGTGGIGRET
MAB_4414|M.abscessus_ATCC_1997 LAFAKDIRRSTRAQAAL-SWRHFESESIEGT-----PVGVLGTGPIGQAI
: : * ::* * :.
TH_3383|M.thermoresistible__bu EQIVAAMGAEVLHTS------TADDGH-PNWRPLNDLLARSDIVSIHVPL
Mvan_0404|M.vanbaalenii_PYR-1 ERILVAMGADVIHTS------TRDDGS-AGWRSLPDLLSACDIVSLHLPL
Mb3020c|M.bovis_AF2122/97 AQRIAAFGAYVVAYDPYVSPARAAQLG-IELLSLDDLLARADFISVHLPK
Rv2996c|M.tuberculosis_H37Rv AQRIAAFGAYVVAYDPYVSPARAAQLG-IELLSLDDLLARADFISVHLPK
MLBr_01692|M.leprae_Br4923 AARIAAFGAHVIAYDPYVAPARAAQLG-IELMSFDDLLARADFISVHLPK
Mflv_4768|M.gilvum_PYR-GCK AKRGAGFGQEVVAFDPFVK---ASPAG-VSMVELDELLATSRFVVLACPL
MMAR_5282|M.marinum_M TEYAKAFGMDVIAWSQNLTEEAAAAHG-VRRVDKPVLFAQSDVVSIHVVL
MUL_4355|M.ulcerans_Agy99 TEYAKAFGMDVIAWSQNLTEEAAAAHG-VRRVDKPVLFAQSDMVSIHVVL
MAV_0349|M.avium_104 AGFAAAFGMEVIAWSQNLTDDAAAAVG-ARRVSKAALFESADVVSVHLVL
MSMEG_2496|M.smegmatis_MC2_155 ARLLRAAGLEVRGAGRTARDDDPDFGRVVASSDLSAEVGWCDHLILAVPL
MAB_4414|M.abscessus_ATCC_1997 STLLGAVGMRVTVLGR------------AESADLSSHVGEFEYLVLAAPL
. * * . . : :
TH_3383|M.thermoresistible__bu TEKTLGLLDATALSRMKPAAVLVNTARGAIVDEIALTEALHARRLAAAGL
Mvan_0404|M.vanbaalenii_PYR-1 TDATAGLLGRDALARMKSDAVLVNTSRGPIVDEEALADALRTGKLAAAGL
Mb3020c|M.bovis_AF2122/97 TPETAGLIDKEALAKTKPGVIIVNAARGGLVDEAALADAITGGHVRAAGL
Rv2996c|M.tuberculosis_H37Rv TPETAGLIDKEALAKTKPGVIIVNAARGGLVDEAALADAITGGHVRAAGL
MLBr_01692|M.leprae_Br4923 TPETAGLIDKEALAKTKPGVIIVNAARGGLVDEVALADAVRSGHVRAAGL
Mflv_4768|M.gilvum_PYR-GCK TPETFHLIDADRLAGMRSDSFLVNVARGPVVLEAALIDALKAGRLAGAAL
MMAR_5282|M.marinum_M SERTRALVGAGELAAMKPGAYLINTSRGPIVDEGALIGALEAGRIAGAGL
MUL_4355|M.ulcerans_Agy99 SERTRALVGAGELAAMKPGAYLINTSRGPIVDEGALIGALEAGRIAGAGL
MAV_0349|M.avium_104 SERTRGLVGEPELALMKPSAYLINTARGPIVDEPALIAALTAGRIAGAGL
MSMEG_2496|M.smegmatis_MC2_155 TDATRGLVDAAVLGAMKPDAHLVNIARGPVVDEAALIAALTEHRIGGATL
MAB_4414|M.abscessus_ATCC_1997 TPQTRGIVNARVLSAMRPTARLINVGRGELVGTWDLVSALNRGGIAGAAL
: * ::. *. :. ::* .** :* * *: : .* *
TH_3383|M.thermoresistible__bu DVFATEPVRPNNPLLTLDNVVLTPHVSWYTVDTMRRYLTHAVDNCRRLRD
Mvan_0404|M.vanbaalenii_PYR-1 DVFAVEPVPADNPLLRLPNVVLTPHVTWYTADTMRRYLEFAVDNCERLRD
Mb3020c|M.bovis_AF2122/97 DVFATEPC-TDSPLFELAQVVVTPHLGASTAEAQDRAGTDVAESVRLALA
Rv2996c|M.tuberculosis_H37Rv DVFATEPC-TDSPLFELAQVVVTPHLGASTAEAQDRAGTDVAESVRLALA
MLBr_01692|M.leprae_Br4923 DVFATEPC-TDSPLFELSQVVVTPHLGASTAEAQDRAGTDVAESVRLALA
Mflv_4768|M.gilvum_PYR-GCK DVFEVEPLPLDSELRTLPNVVLGAHNGSNTREGVVRASNAAVEFLIEELA
MMAR_5282|M.marinum_M DVYDVEPLPVDHRLRSLPNVTLSPHLGYVTREMLSACYADTVEAVVAWLD
MUL_4355|M.ulcerans_Agy99 DVYDVEPLPVDHRLRSLPNVTLSPHLGYVTREMLSVCYADTVEAVVAWLD
MAV_0349|M.avium_104 DVYDVEPLPGDHPLRSLPNVTLSPHLGYVTREMLGAFYADTVESVLAWLD
MSMEG_2496|M.smegmatis_MC2_155 DAFDVEPLPADHPLWDVPNVTITAHMSGDVVGWRDTLAEQYAENVRRWLA
MAB_4414|M.abscessus_ATCC_1997 DVTDPEPLPVGHPLWRTPNTHITPHNSGDVRGWSDRLQDQFVVNFERYLR
*. ** . * :. : .* . .
TH_3383|M.thermoresistible__bu GEQLADVVNGITGRRPG---------------------------------
Mvan_0404|M.vanbaalenii_PYR-1 GRNLVNVVNDVAG-------------------------------------
Mb3020c|M.bovis_AF2122/97 GEFVPDAVNVGGGVVNEEVAPWLDLVRKLGVLAGVLSDELPVSLSVQVRG
Rv2996c|M.tuberculosis_H37Rv GEFVPDAVNVGGGVVNEEVAPWLDLVRKLGVLAGVLSDELPVSLSVQVRG
MLBr_01692|M.leprae_Br4923 GEFVPDAVNVDGGVVNEEVAPWLDLVCKLGVLVAALSDELPASLSVHVRG
Mflv_4768|M.gilvum_PYR-GCK R-------------------------------------------------
MMAR_5282|M.marinum_M GAPIRIANPQAVQRPPS---------------------------------
MUL_4355|M.ulcerans_Agy99 GAPIRIANPQAVRRPPS---------------------------------
MAV_0349|M.avium_104 GTPIRIANPEVLQRD-----------------------------------
MSMEG_2496|M.smegmatis_MC2_155 GEPLHNVVDKKLGYISQAGRGVR---------------------------
MAB_4414|M.abscessus_ATCC_1997 GEELLNIVDKNRMHRHA---------------------------------
TH_3383|M.thermoresistible__bu --------------------------------------------------
Mvan_0404|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3020c|M.bovis_AF2122/97 ELAAEEVEVLRLSALRGLFSAVIEDAVTFVNAPALAAERGVTAEICKASE
Rv2996c|M.tuberculosis_H37Rv ELAAEEVEVLRLSALRGLFSAVIEDAVTFVNAPALAAERGVTAEICKASE
MLBr_01692|M.leprae_Br4923 ELASEDVEILRLSALRGLFSTVIEDAVTFVNAPALAAERGVSAEITTGSE
Mflv_4768|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_5282|M.marinum_M --------------------------------------------------
MUL_4355|M.ulcerans_Agy99 --------------------------------------------------
MAV_0349|M.avium_104 --------------------------------------------------
MSMEG_2496|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4414|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3383|M.thermoresistible__bu --------------------------------------------------
Mvan_0404|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3020c|M.bovis_AF2122/97 SPNHRSVVDVRAVGADGSVVTVSGTLYGPQLSQKIVQINGRHFDLRAQGI
Rv2996c|M.tuberculosis_H37Rv SPNHRSVVDVRAVGADGSVVTVSGTLYGPQLSQKIVQINGRHFDLRAQGI
MLBr_01692|M.leprae_Br4923 SPNHRSVVDVRAVASDGSVVNIAGTLSGPQLVQKIVQVNGRNFDLRAQGM
Mflv_4768|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_5282|M.marinum_M --------------------------------------------------
MUL_4355|M.ulcerans_Agy99 --------------------------------------------------
MAV_0349|M.avium_104 --------------------------------------------------
MSMEG_2496|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4414|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3383|M.thermoresistible__bu --------------------------------------------------
Mvan_0404|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3020c|M.bovis_AF2122/97 NLIIHYVDRPGALGKIGTLLGTAGVNIQAAQLSEDAEGPGATILLRLDQD
Rv2996c|M.tuberculosis_H37Rv NLIIHYVDRPGALGKIGTLLGTAGVNIQAAQLSEDAEGPGATILLRLDQD
MLBr_01692|M.leprae_Br4923 NLVIRYVDQPGALGKIGTLLGAAGVNIQAAQLSEDTEGPGATILLRLDQD
Mflv_4768|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_5282|M.marinum_M --------------------------------------------------
MUL_4355|M.ulcerans_Agy99 --------------------------------------------------
MAV_0349|M.avium_104 --------------------------------------------------
MSMEG_2496|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4414|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3383|M.thermoresistible__bu ------------------------
Mvan_0404|M.vanbaalenii_PYR-1 ------------------------
Mb3020c|M.bovis_AF2122/97 VPDDVRTAIAAAVDAYKLEVVDLS
Rv2996c|M.tuberculosis_H37Rv VPDDVRTAIAAAVDAYKLEVVDLS
MLBr_01692|M.leprae_Br4923 VPGDVRSAIVAAVSANKLEVVNLS
Mflv_4768|M.gilvum_PYR-GCK ------------------------
MMAR_5282|M.marinum_M ------------------------
MUL_4355|M.ulcerans_Agy99 ------------------------
MAV_0349|M.avium_104 ------------------------
MSMEG_2496|M.smegmatis_MC2_155 ------------------------
MAB_4414|M.abscessus_ATCC_1997 ------------------------