For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRVLAHFLPGPKVTEFLSPHTDWLDIRYCAEDDDETFYRELPDAEVIWHVLRPISGDDLSKAPKLRLVHK LGAGVNTVDVDTATARGIAVANMPGANAPSVAEGTLLLMLAALRRLPELDRATRTGNGWPSDPTLGESVR DLGGCTVGLVGYGNIAKQVERILLAIGAEVVHTSTRDDGLPGWRTLPDLLSESDIVSLHLPLTGATEGLL GRKALAQMKTDAVLVNTSRGAIVDEAALVDALRAGTLAAAGLDVFAVEPVPSDNPLLGLPNVVLTPHVTW YTADTMRRYLEFALDNCERLRDGRALAHVVNDPSG
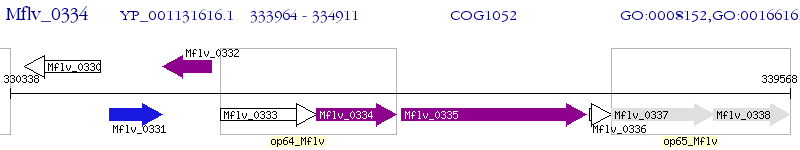
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0334 | - | - | 100% (315) | D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding |
| M. gilvum PYR-GCK | Mflv_4234 | - | 4e-44 | 38.18% (275) | D-3-phosphoglycerate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4768 | - | 2e-31 | 38.63% (233) | D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3020c | serA1 | 3e-45 | 38.57% (293) | D-3-phosphoglycerate dehydrogenase |
| M. tuberculosis H37Rv | Rv2996c | serA1 | 3e-45 | 38.57% (293) | D-3-phosphoglycerate dehydrogenase |
| M. leprae Br4923 | MLBr_01692 | serA | 2e-43 | 38.32% (274) | D-3-phosphoglycerate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4334 | - | 1e-128 | 71.01% (307) | 2-hydroxyacid dehydrogenase |
| M. marinum M | MMAR_0531 | serA3 | 1e-135 | 75.56% (311) | D-3-phosphoglycerate dehydrogenase SerA3 |
| M. avium 104 | MAV_4438 | - | 1e-44 | 43.51% (239) | 2-hydroxyacid dehydrogenase family protein |
| M. smegmatis MC2 155 | MSMEG_0604 | - | 1e-140 | 75.40% (313) | glyoxylate reductase |
| M. thermoresistible (build 8) | TH_3383 | - | 1e-132 | 72.70% (315) | PUTATIVE D-isomer specific 2-hydroxyacid dehydrogenase, |
| M. ulcerans Agy99 | MUL_1195 | serA3 | 1e-134 | 75.24% (311) | D-3-phosphoglycerate dehydrogenase SerA3 |
| M. vanbaalenii PYR-1 | Mvan_0404 | - | 1e-153 | 83.02% (318) | D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3020c|M.bovis_AF2122/97 --------------------------------MSLPVVLIAD---KLAPS
Rv2996c|M.tuberculosis_H37Rv --------------------------------MSLPVVLIAD---KLAPS
MLBr_01692|M.leprae_Br4923 --------------------------------MDLPVVLIAD---KLAQS
Mflv_0334|M.gilvum_PYR-GCK --------------------------------MRVLAHFLPG---PKVTE
Mvan_0404|M.vanbaalenii_PYR-1 --------------------------------MRVLAHFVPGGPDSKVIE
MSMEG_0604|M.smegmatis_MC2_155 -----------------------MAVN--PGTVRVLAHFAPS---NTVLD
MMAR_0531|M.marinum_M -----------------------MN-----QPLRVLAHFVPG---EKVLE
MUL_1195|M.ulcerans_Agy99 -----------------------MN-----QPLRVLAHFVPG---EKVLE
TH_3383|M.thermoresistible__bu VDRPAVALSANRRRHCPGMLRGDMSGVNSRSALRVLAHFRPG---PKVLE
MAB_4334|M.abscessus_ATCC_1997 -----------------------MSLSGENHRVRVLAHFPSG---PRVLE
MAV_4438|M.avium_104 -------------------------------MTSKPRALVTAPLRGPGLD
: . .
Mb3020c|M.bovis_AF2122/97 TVAALGDQVEVRWVDGPDRDKLLAAVPEADALLVRSATTVDAEVLAAAPK
Rv2996c|M.tuberculosis_H37Rv TVAALGDQVEVRWVDGPDRDKLLAAVPEADALLVRSATTVDAEVLAAAPK
MLBr_01692|M.leprae_Br4923 TVAALGDQVEVRWVDGPDRTKLLAAVPEADALLVRSATTVDAEVLAAAPK
Mflv_0334|M.gilvum_PYR-GCK FLSPHTDWLDIRYCAEDDDETFYRELPDAEVIWH-VLRPISGDDLSKAPK
Mvan_0404|M.vanbaalenii_PYR-1 FLAPCRDWLDVRFCAEDDDDTFYRELADAEVIWH-VLRPLSGEDLARAPR
MSMEG_0604|M.smegmatis_MC2_155 FLAPHLDWLDVRFCGEEDDETFYRELGDADVLWH-VLRPITGDDLNRAPR
MMAR_0531|M.marinum_M FVAAQRDWLDIRWCAEDDDSGFYRELDHAEVIWH-VLRPLSGADLQRAPR
MUL_1195|M.ulcerans_Agy99 FVAAQRDWLDIRWCAEDDDSGFYRELDHAEVIWH-VLRPLSGADLQRAPR
TH_3383|M.thermoresistible__bu FLEPESGWLDVRFCDEDDDETFHRELPHAEVIWH-VLRPLSGEDLTRATR
MAB_4334|M.abscessus_ATCC_1997 QLAPHADWLDVRFCAEDDDNTFYAELPGADVLWH-VLRPLSADDVAKGER
MAV_4438|M.avium_104 KLRALAEVVYDPWIEQRPLRIYSAEQLAERIAAEEAEIVVVESDSVRGPV
: . : : : .
Mb3020c|M.bovis_AF2122/97 LKIVARAGVG----LDNVDVDAATARGVLVVNAPTSNIHSAAEHALALLL
Rv2996c|M.tuberculosis_H37Rv LKIVARAGVG----LDNVDVDAATARGVLVVNAPTSNIHSAAEHALALLL
MLBr_01692|M.leprae_Br4923 LKIVARAGVG----LDNVDVDAATARGVLVVNAPTSNIHSAAEHALALLL
Mflv_0334|M.gilvum_PYR-GCK LRLVHKLGAG----VNTVDVDTATARGIAVANMPGANAPSVAEGTLLLML
Mvan_0404|M.vanbaalenii_PYR-1 LRLVHKLGAG----VNTIDVDAATARGVAVANMPGANAESVAEGTLLLML
MSMEG_0604|M.smegmatis_MC2_155 LRLVHKLGAG----VNTIDVETATQLGILVANMPGANAPSVAEGTVLLML
MMAR_0531|M.marinum_M LRLVQKLGAG----VNTIDVATATKHGIAVANMPGANAPSVAEGTVLLML
MUL_1195|M.ulcerans_Agy99 LRLVQKLGAG----VNTIDVATATKHGIAVANMPGANAPSVAEGTVLLML
TH_3383|M.thermoresistible__bu CRLVHKLGTG----VNTIDVDTATERGIAVANMPGANAASVAEGTVMLML
MAB_4334|M.abscessus_ATCC_1997 LRLIHKFGAG----VNTIALDAAVEHGVAVANMPGANAPSVAEGALLLML
MAV_4438|M.avium_104 FELGVRAIAATRGDPNNVDIDGATAAGIPVLNTPGRNADAVAEMTVALLL
.: : .. :.: : *. *: * * * * :.** :: *:*
Mb3020c|M.bovis_AF2122/97 AASRQIPAADASLREHTWKRSS--FSG----TEIFGKTVGVVGLGRIGQL
Rv2996c|M.tuberculosis_H37Rv AASRQIPAADASLREHTWKRSS--FSG----TEIFGKTVGVVGLGRIGQL
MLBr_01692|M.leprae_Br4923 AASRQIAEADASLRAHIWKRSS--FSG----TEIFGKTVGVVGLGRIGQL
Mflv_0334|M.gilvum_PYR-GCK AALRRLPELDRATRTGNGWPSD--PTLGESVRDLGGCTVGLVGYGNIAKQ
Mvan_0404|M.vanbaalenii_PYR-1 AALRRLPVLDRATREGRGWPSD--PTLGETVRDIGGCTVGLVGYGNIAKQ
MSMEG_0604|M.smegmatis_MC2_155 AALRRLPQLDRATRAGRGWPTD--PTLGDTVRDIGGCTVGLVGYGNVAKR
MMAR_0531|M.marinum_M AALRRLPELDAATRQGRGWPTD--PQLGESVRDIGSCTVGLIGYGNIAKR
MUL_1195|M.ulcerans_Agy99 AALRRLPELDAATRQGRGWPTD--PQLGESVRDIGSCTVGLIGYGNIAKR
TH_3383|M.thermoresistible__bu AALRRLVELDRATRAGTGWPTD--PSLGETVRDIGGSTVGLIGYGNVARR
MAB_4334|M.abscessus_ATCC_1997 AALRQLPRLDRDIRSGNGWPTD--QSLGESVRDIGSCTIGLVGYGNIAKS
MAV_4438|M.avium_104 AATRHLLPADADVRGGNIFRDGSIPYQRFRGGEIAGLTAGLVGLGAVGRA
** *:: * * . :: . * *::* * :.:
Mb3020c|M.bovis_AF2122/97 VAQRIAAFGAYVVAYDPYVSPARAAQLGIELLSLDDLLARADFISVHLPK
Rv2996c|M.tuberculosis_H37Rv VAQRIAAFGAYVVAYDPYVSPARAAQLGIELLSLDDLLARADFISVHLPK
MLBr_01692|M.leprae_Br4923 VAARIAAFGAHVIAYDPYVAPARAAQLGIELMSFDDLLARADFISVHLPK
Mflv_0334|M.gilvum_PYR-GCK VERILLAIGAE-VVHTSTRDDGLPGWR-----TLPDLLSESDIVSLHLPL
Mvan_0404|M.vanbaalenii_PYR-1 VERILVAMGAD-VIHTSTRDDGSAGWR-----SLPDLLSACDIVSLHLPL
MSMEG_0604|M.smegmatis_MC2_155 VERIVLAMGAEQVLHTSTRDTGHPRWR-----NLPDLLAASDIVSLHLPL
MMAR_0531|M.marinum_M VATIVSAMGAT-VLHTSTRDDGRPGWR-----PLPELLATSDIVSLHLPL
MUL_1195|M.ulcerans_Agy99 VATIVSAMGAT-VLHTSTRDDGRPGWR-----PLPELLATSDIVSLHLPL
TH_3383|M.thermoresistible__bu VEQIVAAMGAE-VLHTSTADDGHPNWR-----PLNDLLARSDIVSIHVPL
MAB_4334|M.abscessus_ATCC_1997 LEKILLAMGAT-VLHTSTRDDGTAGWR-----GLNDLLTSSDIVSLHLPL
MAV_4438|M.avium_104 TRWRLSGLGLRVIAHDPYHPEAR--------HGLDELLAESDVVSLHAPV
: .:* : : . . : :**: .*.:*:* *
Mb3020c|M.bovis_AF2122/97 TPETAGLIDKEALAKTKPGVIIVNAARGGLVDEAALADAITGGHVRAAGL
Rv2996c|M.tuberculosis_H37Rv TPETAGLIDKEALAKTKPGVIIVNAARGGLVDEAALADAITGGHVRAAGL
MLBr_01692|M.leprae_Br4923 TPETAGLIDKEALAKTKPGVIIVNAARGGLVDEVALADAVRSGHVRAAGL
Mflv_0334|M.gilvum_PYR-GCK TGATEGLLGRKALAQMKTDAVLVNTSRGAIVDEAALVDALRAGTLAAAGL
Mvan_0404|M.vanbaalenii_PYR-1 TDATAGLLGRDALARMKSDAVLVNTSRGPIVDEEALADALRTGKLAAAGL
MSMEG_0604|M.smegmatis_MC2_155 TDTSRGLLGPEAIAAMKPGAVLVNTARGPIVDEAALIEALRGGRLAAAGL
MMAR_0531|M.marinum_M TPHTDRLLDADALAQMKPNSLLINTSRGAVIDEDALVDALRSGSLAAAGL
MUL_1195|M.ulcerans_Agy99 TPHTDRLLDADALARMKPNSLLINTSRGAVVDEDALVDALRSGSLAAAGL
TH_3383|M.thermoresistible__bu TEKTLGLLDATALSRMKPAAVLVNTARGAIVDEIALTEALHARRLAAAGL
MAB_4334|M.abscessus_ATCC_1997 TEASTGMLDSAALNRMKPGSVLVNTSRGAVVDEAALVNALQQGPLGAAGL
MAV_4438|M.avium_104 TDETTGMIGAEQFAAMRDGVVFLNTARAQLHDTDALVEALRAGKVAAAGL
* : ::. : : :::*::*. : * ** :*: : ****
Mb3020c|M.bovis_AF2122/97 DVFATEPCT-DSPLFELAQVVVTPHLGASTAEAQDRAGTDVAESVRLALA
Rv2996c|M.tuberculosis_H37Rv DVFATEPCT-DSPLFELAQVVVTPHLGASTAEAQDRAGTDVAESVRLALA
MLBr_01692|M.leprae_Br4923 DVFATEPCT-DSPLFELSQVVVTPHLGASTAEAQDRAGTDVAESVRLALA
Mflv_0334|M.gilvum_PYR-GCK DVFAVEPVPSDNPLLGLPNVVLTPHVTWYTADTMRRYLEFALDNCERLRD
Mvan_0404|M.vanbaalenii_PYR-1 DVFAVEPVPADNPLLRLPNVVLTPHVTWYTADTMRRYLEFAVDNCERLRD
MSMEG_0604|M.smegmatis_MC2_155 DVFDTEPLPADHPLLGLDNVVLTPHVTWYTADTMRRYLSIGVENCRRIRD
MMAR_0531|M.marinum_M DVFAVEPVVPENPLLRLDNVVLTPHVTWYTVDTMRRYLTEAVDNCRRLRD
MUL_1195|M.ulcerans_Agy99 DVFAVEPVVPENPLLRLDNVVLTPHVTWYTVDTMRRYLTEAVDNCRRLRD
TH_3383|M.thermoresistible__bu DVFATEPVRPNNPLLTLDNVVLTPHVSWYTVDTMRRYLTHAVDNCRRLRD
MAB_4334|M.abscessus_ATCC_1997 DVFAQEPVSPENPLLALPNVVLTPHVTWFTADTMTRYLHHAIDNCRRIHE
MAV_4438|M.avium_104 DHFVGEWLPTDHPLVGMPNVVLTPHIGGATWNTEARQAQLVADDLEALLA
* * * : **. : :**:***: * :: * :. .
Mb3020c|M.bovis_AF2122/97 GEFVPDAVNVGGGVVNEEVAPWLDLVRKLGVLAGVLSDELPVSLSVQVRG
Rv2996c|M.tuberculosis_H37Rv GEFVPDAVNVGGGVVNEEVAPWLDLVRKLGVLAGVLSDELPVSLSVQVRG
MLBr_01692|M.leprae_Br4923 GEFVPDAVNVDGGVVNEEVAPWLDLVCKLGVLVAALSDELPASLSVHVRG
Mflv_0334|M.gilvum_PYR-GCK GRALAHVVNDPSG-------------------------------------
Mvan_0404|M.vanbaalenii_PYR-1 GRNLVNVVNDVAG-------------------------------------
MSMEG_0604|M.smegmatis_MC2_155 GEPLAHLVNEAR--------------------------------------
MMAR_0531|M.marinum_M GLSLANVVNCQADC------------------------------------
MUL_1195|M.ulcerans_Agy99 GLSLANAVNCQADC------------------------------------
TH_3383|M.thermoresistible__bu GEQLADVVNGITGRRPG---------------------------------
MAB_4334|M.abscessus_ATCC_1997 GMPLADRVR-----------------------------------------
MAV_4438|M.avium_104 GATPAHIVNPEVLAR-----------------------------------
* . *.
Mb3020c|M.bovis_AF2122/97 ELAAEEVEVLRLSALRGLFSAVIEDAVTFVNAPALAAERGVTAEICKASE
Rv2996c|M.tuberculosis_H37Rv ELAAEEVEVLRLSALRGLFSAVIEDAVTFVNAPALAAERGVTAEICKASE
MLBr_01692|M.leprae_Br4923 ELASEDVEILRLSALRGLFSTVIEDAVTFVNAPALAAERGVSAEITTGSE
Mflv_0334|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0404|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0604|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_0531|M.marinum_M --------------------------------------------------
MUL_1195|M.ulcerans_Agy99 --------------------------------------------------
TH_3383|M.thermoresistible__bu --------------------------------------------------
MAB_4334|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_4438|M.avium_104 --------------------------------------------------
Mb3020c|M.bovis_AF2122/97 SPNHRSVVDVRAVGADGSVVTVSGTLYGPQLSQKIVQINGRHFDLRAQGI
Rv2996c|M.tuberculosis_H37Rv SPNHRSVVDVRAVGADGSVVTVSGTLYGPQLSQKIVQINGRHFDLRAQGI
MLBr_01692|M.leprae_Br4923 SPNHRSVVDVRAVASDGSVVNIAGTLSGPQLVQKIVQVNGRNFDLRAQGM
Mflv_0334|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0404|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0604|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_0531|M.marinum_M --------------------------------------------------
MUL_1195|M.ulcerans_Agy99 --------------------------------------------------
TH_3383|M.thermoresistible__bu --------------------------------------------------
MAB_4334|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_4438|M.avium_104 --------------------------------------------------
Mb3020c|M.bovis_AF2122/97 NLIIHYVDRPGALGKIGTLLGTAGVNIQAAQLSEDAEGPGATILLRLDQD
Rv2996c|M.tuberculosis_H37Rv NLIIHYVDRPGALGKIGTLLGTAGVNIQAAQLSEDAEGPGATILLRLDQD
MLBr_01692|M.leprae_Br4923 NLVIRYVDQPGALGKIGTLLGAAGVNIQAAQLSEDTEGPGATILLRLDQD
Mflv_0334|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0404|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0604|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_0531|M.marinum_M --------------------------------------------------
MUL_1195|M.ulcerans_Agy99 --------------------------------------------------
TH_3383|M.thermoresistible__bu --------------------------------------------------
MAB_4334|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_4438|M.avium_104 --------------------------------------------------
Mb3020c|M.bovis_AF2122/97 VPDDVRTAIAAAVDAYKLEVVDLS
Rv2996c|M.tuberculosis_H37Rv VPDDVRTAIAAAVDAYKLEVVDLS
MLBr_01692|M.leprae_Br4923 VPGDVRSAIVAAVSANKLEVVNLS
Mflv_0334|M.gilvum_PYR-GCK ------------------------
Mvan_0404|M.vanbaalenii_PYR-1 ------------------------
MSMEG_0604|M.smegmatis_MC2_155 ------------------------
MMAR_0531|M.marinum_M ------------------------
MUL_1195|M.ulcerans_Agy99 ------------------------
TH_3383|M.thermoresistible__bu ------------------------
MAB_4334|M.abscessus_ATCC_1997 ------------------------
MAV_4438|M.avium_104 ------------------------