For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MIEIENLTKRFGDRTVLDEVSLSVAAGEILAVVGPSGAGKSTLSRCVSALERPTSGTVRVDGKDFSRLDG TELIAARRGVGVIFQSAPLLRRRTVARNIALPLEYLGATQSSIDKRVSELLERVGLTDRASYLPAQLSGG QKQRVGIARALALGPSNLLSDEATSGLDPATTKSILELLSHLRDEYALSIILITHEMEVVREIADSVARL DGGRIIESGRVDDIILDPDSTLARELLPDRPGALAPGAGEVWEVSYASREVPLDWLTSIQSAPGLGDAKI SVLSASIEAIRGVAVGRATLSIEPAAPTRLRRAPA
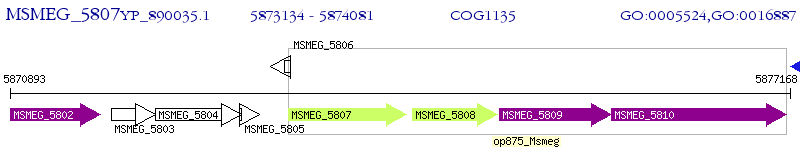
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5807 | - | - | 100% (315) | D-methionine transport ATP-binding protein MetN |
| M. smegmatis MC2 155 | MSMEG_3056 | - | 1e-57 | 51.67% (240) | ABC transporter ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_5371 | ehuA | 2e-46 | 41.30% (247) | ectoine/hydroxyectoine ABC transporter, ATP-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3129c | ftsE | 1e-39 | 38.53% (218) | putative cell division ATP-binding protein FTSE (septation |
| M. gilvum PYR-GCK | Mflv_3165 | - | 3e-43 | 42.19% (237) | ABC transporter related |
| M. tuberculosis H37Rv | Rv3102c | ftsE | 1e-39 | 38.53% (218) | putative cell division ATP-binding protein FTSE (septation |
| M. leprae Br4923 | MLBr_00669 | ftsE | 2e-39 | 38.07% (218) | putative cell division ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_2803c | - | 5e-62 | 43.85% (317) | methionine import ATP-binding protein metN |
| M. marinum M | MMAR_1530 | ftsE | 4e-39 | 38.07% (218) | cell division ATP-binding protein FtsE |
| M. avium 104 | MAV_4003 | ftsE | 5e-39 | 37.61% (218) | cell division ATP-binding protein FtsE |
| M. thermoresistible (build 8) | TH_2538 | - | 7e-77 | 60.71% (252) | PUTATIVE ABC transporter related |
| M. ulcerans Agy99 | MUL_2409 | ftsE | 3e-39 | 38.07% (218) | cell division ATP-binding protein FtsE |
| M. vanbaalenii PYR-1 | Mvan_3884 | - | 1e-138 | 81.49% (308) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00669|M.leprae_Br4923 -----------------MITLDHVSKKYKSLAR--PALDNVNVKIDKGEF
MAV_4003|M.avium_104 ----------------MMITLDHVTKKYKASAR--PALEDVNVKIDKGEF
Mb3129c|M.bovis_AF2122/97 -----------------MITLDHVTKQYKSSAR--PALDDINVKIDKGEF
Rv3102c|M.tuberculosis_H37Rv -----------------MITLDHVTKQYKSSAR--PALDDINVKIDKGEF
MMAR_1530|M.marinum_M -----------------MITLDHVSKQYKSSAR--PALDDINVKIDKGEF
MUL_2409|M.ulcerans_Agy99 -----------------MITLDHVSKQYKSSAR--PALDDINVKIDKGEF
MAB_2803c|M.abscessus_ATCC_199 -----------MGDVAPLIQFQNVTKTFRVGKKTVTAVDDVSLDIGSGDV
MSMEG_5807|M.smegmatis_MC2_155 -----------------MIEIENLTKRFG----DRTVLDEVSLSVAAGEI
Mvan_3884|M.vanbaalenii_PYR-1 --MLRVTAGPSTVESFPVIEIENLTKRFG----DRTVLDDISLSVGSGEI
TH_2538|M.thermoresistible__bu -----------------MIELTDLTKVYGCGEHRTVVLDRINFRVETGEV
Mflv_3165|M.gilvum_PYR-GCK MSELITEAAASEPAGTVKIRIEGLKKSFG----DLVVLDGIDTTVSHGEV
* : :.* : .:: :. : *:.
MLBr_00669|M.leprae_Br4923 VFLIGPSGSGKSTFMRLLLGAETPTSGDVRVSKFHVNKLPGRHIPRLRQV
MAV_4003|M.avium_104 VFLIGPSGSGKSTFMRLLLGEENPTTGDIRVSKFHVNKLRGRHIPKLRQV
Mb3129c|M.bovis_AF2122/97 VFLIGPSGSGKSTFMRLLLAAETPTSGDVRVSKFHVNKLRGRHVPKLRQV
Rv3102c|M.tuberculosis_H37Rv VFLIGPSGSGKSTFMRLLLAAETPTSGDVRVSKFHVNKLRGRHVPKLRQV
MMAR_1530|M.marinum_M VFLIGPSGSGKSTFMRLLLAAETPTSGDVRVSKFHVNKLRGRNVPKLRQV
MUL_2409|M.ulcerans_Agy99 VFLIGPSGSGKSTFMRLLLAAETPTSGDVRVSKFHVNKLRGRNVPKLRQV
MAB_2803c|M.abscessus_ATCC_199 FAMIGYSGAGKSTLVRLINGLERPTSGRVIVDGAEVSALGERELRGLRSD
MSMEG_5807|M.smegmatis_MC2_155 LAVVGPSGAGKSTLSRCVSALERPTSGTVRVDGKDFSRLDGTELIAARRG
Mvan_3884|M.vanbaalenii_PYR-1 LAVVGPSGAGKSTLSRCVSFLERPTSGTVRVDGKDFTRLDGDELLAARRN
TH_2538|M.thermoresistible__bu LAVVGPSGAGKSTLAQCINLLTRPTSGSVVVNGEDLTTLSTHRLRVARRR
Mflv_3165|M.gilvum_PYR-GCK VCVIGPSGSGKSTFLRCLNKLEDITGGRVVVDEFDLTDP-KIDLDKVRQH
. ::* **:****: : : * * : *. ... : *
MLBr_00669|M.leprae_Br4923 IGCVFQDFRLLQQKTVYENVAFA-LEVIGRRSDAINQVVPDVLETVGLSG
MAV_4003|M.avium_104 IGTVFQDFRLLQQKTVYENVAFA-LEVIGKRTDAINRVVPEVLETVGLSG
Mb3129c|M.bovis_AF2122/97 IGCVFQDFRLLQQKTVYDNVAFA-LEVIGKRTDAINRVVPEVLETVGLSG
Rv3102c|M.tuberculosis_H37Rv IGCVFQDFRLLQQKTVYDNVAFA-LEVIGKRTDAINRVVPEVLETVGLSG
MMAR_1530|M.marinum_M IGCVFQDFRLLQQKTVYENVAFA-LEVIGKRADAINRVVPDVLETVGLSG
MUL_2409|M.ulcerans_Agy99 IGCVFQDFRLLQQKTVYENVAFA-LEVIGKRADAINRVVPDVLETVGLSG
MAB_2803c|M.abscessus_ATCC_199 IGMIFQQFNLFRSRTVAGNIAYP-LRVAGWTADKRNARVAELLEFVGLGD
MSMEG_5807|M.smegmatis_MC2_155 VGVIFQSAPLLRRRTVARNIALP-LEYLGATQSSIDKRVSELLERVGLTD
Mvan_3884|M.vanbaalenii_PYR-1 VGVIFQTAPLLRRRTVAQNIALP-LQYLHATEGSAGTRVTELLDRVGLAD
TH_2538|M.thermoresistible__bu IGTVFQSSGLLERRTAAENVALP-LEYLGVTAAETRKRVAELLDRVGLAA
Mflv_3165|M.gilvum_PYR-GCK IGMVFQHFNLFPHMTVLENVTLAPLLTKKMDKAAAEKKALQLLDQVGLAE
:* :** *: *. *:: . * . ::*: ***
MLBr_00669|M.leprae_Br4923 KANRLPDELSGGEQQRVAIARAFVNRPLVLLADEPTGNLDPDTSKDIMDL
MAV_4003|M.avium_104 KANRLPHELSGGEQQRVAIARAFVNRPLVLLADEPTGNLDPDTSKDIMDL
Mb3129c|M.bovis_AF2122/97 KANRLPDELSGGEQQRVAIARAFVNRPLVLLADEPTGNLDPETSRDIMDL
Rv3102c|M.tuberculosis_H37Rv KANRLPDELSGGEQQRVAIARAFVNRPLVLLADEPTGNLDPETSRDIMDL
MMAR_1530|M.marinum_M KANRLPHELSGGEQQRVAIARAFVNRPLVLLADEPTGNLDPDTSKDIMDL
MUL_2409|M.ulcerans_Agy99 KANRLPHELSGGEQQRVAIARAFVNRPLVLLADEPTGNLDPDTSKDIMDL
MAB_2803c|M.abscessus_ATCC_199 KAKSYPDQLSGGQKQRVGIARALATSPSVLLADEATSALDPETTSDVLRL
MSMEG_5807|M.smegmatis_MC2_155 RASYLPAQLSGGQKQRVGIARALALGPSNLLSDEATSGLDPATTKSILEL
Mvan_3884|M.vanbaalenii_PYR-1 RRDFYPAQLSGGQKQRVGIARALALGPSNLLSDEATSGLDPATTRSILAL
TH_2538|M.thermoresistible__bu RANHYPFQLSGGQRQRVGIARALALRPSVLLSDEATAGLDPTTTASVVEL
Mflv_3165|M.gilvum_PYR-GCK KAGVKPATLSGGQKQRVAIARALAMNPSIMLFDEATSALDPEMVGDVLEV
: * ****::***.****:. * :* **.*. *** .:: :
MLBr_00669|M.leprae_Br4923 LERIN-RTGTTVLMATHDHHIVDSMRQRVVELSLGRLVRD----------
MAV_4003|M.avium_104 LERIN-RTGTTVLMATHDHHIVDSMRQRVVELSLGRLVRD----------
Mb3129c|M.bovis_AF2122/97 LERIN-RTGTTVLMATHDHHIVDSMRQRVVELSLGRLVRD----------
Rv3102c|M.tuberculosis_H37Rv LERIN-RTGTTVLMATHDHHIVDSMRQRVVELSLGRLVRD----------
MMAR_1530|M.marinum_M LERIN-RTGTTVLMATHDHHIVDSMRQRVVELSLGRLVRD----------
MUL_2409|M.ulcerans_Agy99 LERIN-RTGTTVLMATHDHHIVDSMRQRVVELSLGRLVRD----------
MAB_2803c|M.abscessus_ATCC_199 LREVNAEFGVTIVVITHEMDVVRTVADHVAVLADGKVVETGTVFDVFSTP
MSMEG_5807|M.smegmatis_MC2_155 LSHLRDEYALSIILITHEMEVVREIADSVARLDGGRIIESGRVDDIILDP
Mvan_3884|M.vanbaalenii_PYR-1 LSHLRDEFGLSIILITHEMEVVREIADSVARIEDGRIIESGSVQDVILDP
TH_2538|M.thermoresistible__bu LRELRDDLNLSIVFITHEMDTVLKIADSVARLDHGRIVEAGRLVDLLTDP
Mflv_3165|M.gilvum_PYR-GCK LRTLARG-GMTMVVVTHEMGFAREVASRVIFMADGHIVEEGPPAEIFDNP
* : :::. **: . : . * : *:::.
MLBr_00669|M.leprae_Br4923 ---------------------------EWCG-------------------
MAV_4003|M.avium_104 ---------------------------EQRG-------------------
Mb3129c|M.bovis_AF2122/97 ---------------------------EQRG-------------------
Rv3102c|M.tuberculosis_H37Rv ---------------------------EQRG-------------------
MMAR_1530|M.marinum_M ---------------------------EQRG-------------------
MUL_2409|M.ulcerans_Agy99 ---------------------------EQRG-------------------
MAB_2803c|M.abscessus_ATCC_199 QSEAGKRFVGTVLRNTPSGEDIARIAQRHKGRIVAVEISEGVQIGPALAR
MSMEG_5807|M.smegmatis_MC2_155 DS-------------------TLARELLPDRPGALAPGAG--EVWEVS--
Mvan_3884|M.vanbaalenii_PYR-1 AS-------------------ELARELLPDRPSVPLDGAG--EIWEVS--
TH_2538|M.thermoresistible__bu AS-------------------ALGADLRPQGAAAPPAPPXXXRLRGVAGP
Mflv_3165|M.gilvum_PYR-GCK KH-------------------PRLQEFLSKVL------------------
MLBr_00669|M.leprae_Br4923 ----------IYG-----MDR-----------------------------
MAV_4003|M.avium_104 ----------IYG-----MDR-----------------------------
Mb3129c|M.bovis_AF2122/97 ----------VYG-----MDR-----------------------------
Rv3102c|M.tuberculosis_H37Rv ----------VYG-----MDR-----------------------------
MMAR_1530|M.marinum_M ----------VYG-----MDR-----------------------------
MUL_2409|M.ulcerans_Agy99 ----------VYG-----MDR-----------------------------
MAB_2803c|M.abscessus_ATCC_199 ATELGVRFEVVYGGITTLQSRSFGNLTLELEGPDDQVAHVVSDLSAVTTV
MSMEG_5807|M.smegmatis_MC2_155 -YASREVPLDWLTSIQSAPGLGDAKISVLSASIEAIRGVAVGRATLSIEP
Mvan_3884|M.vanbaalenii_PYR-1 -HASRTVPLDWLTSIQSAPGLSGAAVSVLSASVESIRGVAVGRAVLAITP
TH_2538|M.thermoresistible__bu DHPHRTGAVRAAGRTGPAAARRCGGGTAVDAGSDRRVADRAGRRPGRRDD
Mflv_3165|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00669|M.leprae_Br4923 --------------------------------
MAV_4003|M.avium_104 --------------------------------
Mb3129c|M.bovis_AF2122/97 --------------------------------
Rv3102c|M.tuberculosis_H37Rv --------------------------------
MMAR_1530|M.marinum_M --------------------------------
MUL_2409|M.ulcerans_Agy99 --------------------------------
MAB_2803c|M.abscessus_ATCC_199 QRVAR---------------------------
MSMEG_5807|M.smegmatis_MC2_155 AAPT---------RLRRAPA------------
Mvan_3884|M.vanbaalenii_PYR-1 SAPSGFVEYLTERGLHVRPVEKVAAGSAEAAA
TH_2538|M.thermoresistible__bu AVAAP----------RRRPIRP----------
Mflv_3165|M.gilvum_PYR-GCK --------------------------------