For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GDVAPLIQFQNVTKTFRVGKKTVTAVDDVSLDIGSGDVFAMIGYSGAGKSTLVRLINGLERPTSGRVIVD GAEVSALGERELRGLRSDIGMIFQQFNLFRSRTVAGNIAYPLRVAGWTADKRNARVAELLEFVGLGDKAK SYPDQLSGGQKQRVGIARALATSPSVLLADEATSALDPETTSDVLRLLREVNAEFGVTIVVITHEMDVVR TVADHVAVLADGKVVETGTVFDVFSTPQSEAGKRFVGTVLRNTPSGEDIARIAQRHKGRIVAVEISEGVQ IGPALARATELGVRFEVVYGGITTLQSRSFGNLTLELEGPDDQVAHVVSDLSAVTTVQRVAR*
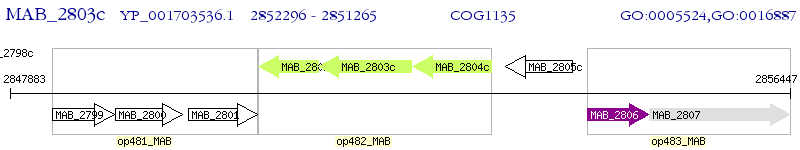
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2803c | - | - | 100% (343) | methionine import ATP-binding protein metN |
| M. abscessus ATCC 19977 | MAB_3050 | - | 4e-48 | 42.51% (247) | putative glutamate ABC transporter, ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_4237c | - | 5e-43 | 42.44% (238) | amino acid ABC transporter ATP-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1312c | oppD | 9e-46 | 39.55% (268) | oligopeptide-transport ATP-binding protein ABC transporter |
| M. gilvum PYR-GCK | Mflv_3968 | - | 2e-51 | 42.97% (249) | ABC transporter related |
| M. tuberculosis H37Rv | Rv1281c | oppD | 9e-46 | 39.55% (268) | oligopeptide-transport ATP-binding protein ABC transporter |
| M. leprae Br4923 | MLBr_00669 | ftsE | 2e-41 | 41.82% (220) | putative cell division ATP-binding protein |
| M. marinum M | MMAR_0060 | - | 2e-43 | 43.50% (223) | glutamine ABC transporter, ATP-binding protein |
| M. avium 104 | MAV_0058 | - | 4e-43 | 45.29% (223) | ABC transporter, ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_3056 | - | 9e-63 | 45.07% (304) | ABC transporter ATP-binding protein |
| M. thermoresistible (build 8) | TH_2538 | - | 3e-58 | 48.95% (237) | PUTATIVE ABC transporter related |
| M. ulcerans Agy99 | MUL_4001 | oppD | 1e-40 | 36.57% (268) | oligopeptide-transport ATP-binding protein ABC transporter |
| M. vanbaalenii PYR-1 | Mvan_3884 | - | 4e-65 | 43.31% (344) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0060|M.marinum_M ----MTRQDAGRQPVCLAARSIHQTLGG----SVVLRGVDLEVPAGTTAA
MAV_0058|M.avium_104 ----MS-NPASRASVSLACKDIHQTLNG----NAVLRGVDLDTPAGSTVA
Mflv_3968|M.gilvum_PYR-GCK ----MP---------MISMKSVNKHFGP----LHVLKDIDLEVGRGQVVV
MAB_2803c|M.abscessus_ATCC_199 ----MG-----DVAPLIQFQNVTKTFRVGKKTVTAVDDVSLDIGSGDVFA
TH_2538|M.thermoresistible__bu ---------------MIELTDLTKVYGCGEHRTVVLDRINFRVETGEVLA
Mvan_3884|M.vanbaalenii_PYR-1 MLRVTAGPSTVESFPVIEIENLTKRFG----DRTVLDDISLSVGSGEILA
MSMEG_3056|M.smegmatis_MC2_155 -------------MMSIYTEDLTKFYG----TTRALEGVSISVDQGEILG
MLBr_00669|M.leprae_Br4923 ---------------MITLDHVSKKYKS--LARPALDNVNVKIDKGEFVF
Mb1312c|M.bovis_AF2122/97 ------------MSPLLEVTDLAVTFRTDGDPVTAVRGISYRVEPGEVVA
Rv1281c|M.tuberculosis_H37Rv ------------MSPLLEVTDLAVTFRTDGDPVTAVRGISYRVEPGEVVA
MUL_4001|M.ulcerans_Agy99 ------------MNPLLEVTDLAVTFPTENDPVTAVRGISYQVAPGEVVA
: : .: :. *
MMAR_0060|M.marinum_M VIGPSGSGKSTLLRTLNRLHEP---DSGDILLDGRSVLRDNP---NDLR-
MAV_0058|M.avium_104 VIGPSGSGKSTLLRTLNRLYEP---DRGDILLDGRSVLHDDP---DQLR-
Mflv_3968|M.gilvum_PYR-GCK VLGPSGSGKSTLCRTINRLETI---DSGTIAIDGVELPAEGRK-LAQLR-
MAB_2803c|M.abscessus_ATCC_199 MIGYSGAGKSTLVRLINGLERP---TSGRVIVDGAEVSALGERELRGLR-
TH_2538|M.thermoresistible__bu VVGPSGAGKSTLAQCINLLTRP---TSGSVVVNGEDLTTLSTHRLRVAR-
Mvan_3884|M.vanbaalenii_PYR-1 VVGPSGAGKSTLSRCVSFLERP---TSGTVRVDGKDFTRLDGDELLAAR-
MSMEG_3056|M.smegmatis_MC2_155 VVGSSGSGKSTLVRNIALLERP---TRGRVVLDGQDLTALSEKELRGAR-
MLBr_00669|M.leprae_Br4923 LIGPSGSGKSTFMRLLLGAETP---TSGDVRVSKFHVNKLPGRHIPRLR-
Mb1312c|M.bovis_AF2122/97 MVGESGSGKSAAAMAVVGLLPEYAQVRGSVRLQGTELLGLADNAMSRFRG
Rv1281c|M.tuberculosis_H37Rv MVGESGSGKSAAAMAVVGLLPEYAQVRGSVRLQGTELLGLADNAMSRFRG
MUL_4001|M.ulcerans_Agy99 VVGESGSGKSAAAMAVVGLLPEYAKVRGSVRLQGTELLGLADDAMSRFRG
::* **:***: : * : :. . *
MMAR_0060|M.marinum_M QRIGMVFQH-FNLFPHRNVVDNVTLAPRKLRG--LSNEEARMLALDQLDR
MAV_0058|M.avium_104 QRIGMVFQH-FNLFPHRTVLDNVALAPRKLRR--LPAEAARELALTQLDR
Mflv_3968|M.gilvum_PYR-GCK SDVGMVFQS-FNLFAHKTILENVTLAPMKVRK--FSKDESREKAMALLER
MAB_2803c|M.abscessus_ATCC_199 SDIGMIFQQ-FNLFRSRTVAGNIAY-PLRVAG--WTADKRNARVAELLEF
TH_2538|M.thermoresistible__bu RRIGTVFQS-SGLLERRTAAENVAL-PLEYLG--VTAAETRKRVAELLDR
Mvan_3884|M.vanbaalenii_PYR-1 RNVGVIFQT-APLLRRRTVAQNIAL-PLQYLH--ATEGSAGTRVTELLDR
MSMEG_3056|M.smegmatis_MC2_155 RRLGNVFQS-ANLLDNRTARANIEY-PLEIAG--WDRKKRWYRAQELLEL
MLBr_00669|M.leprae_Br4923 QVIGCVFQD-FRLLQQKTVYENVAF-ALEVIG--RRSDAINQVVPDVLET
Mb1312c|M.bovis_AF2122/97 KAIGTVFQDPMSALTPVYTVGDQIAEAIEVHQPRVGKKAARRRAVELLDL
Rv1281c|M.tuberculosis_H37Rv KAIGTVFQDPMSALTPVYTVGDQIAEAIEVHQPRVGKKAARRRAVELLDL
MUL_4001|M.ulcerans_Agy99 KVIGTVFQDPMSALTPVYTVGDQIAETIEIHQPTFSRKAARRRAVELLEL
:* :** : : . . . *:
MMAR_0060|M.marinum_M VGLKH---KANARPDMLSGGQQQRVAIARALAMAPQVMFFDEATSALDPE
MAV_0058|M.avium_104 VGLKH---KAGARPATLSGGQQQRVAIARALAMSPQVMFFDEATSALDPE
Mflv_3968|M.gilvum_PYR-GCK VGVAN---QADKYPAQLSGGQQQRVAIARSLAMNPKVMLFDEPTSALDPE
MAB_2803c|M.abscessus_ATCC_199 VGLGD---KAKSYPDQLSGGQKQRVGIARALATSPSVLLADEATSALDPE
TH_2538|M.thermoresistible__bu VGLAA---RANHYPFQLSGGQRQRVGIARALALRPSVLLSDEATAGLDPT
Mvan_3884|M.vanbaalenii_PYR-1 VGLAD---RRDFYPAQLSGGQKQRVGIARALALGPSNLLSDEATSGLDPA
MSMEG_3056|M.smegmatis_MC2_155 VGLPG---RGDAYPAQLSGGQQQRVGIARALAAHPTVILADEPTSALDPT
MLBr_00669|M.leprae_Br4923 VGLSG---KANRLPDELSGGEQQRVAIARAFVNRPLVLLADEPTGNLDPD
Mb1312c|M.bovis_AF2122/97 VGISQPQRRSRAFPHELSGGERQRVVIAIAIANDPDLLICDEPTTALDVT
Rv1281c|M.tuberculosis_H37Rv VGISQPQRRSRAFPHELSGGERQRVVIAIAIANDPDLLICDEPTTALDVT
MUL_4001|M.ulcerans_Agy99 VGIAQPERRARAFPHELSGGERQRVVIAIAIANDPDLLICDEPTTALDVT
**: : * ****::*** ** ::. * :: **.* **
MMAR_0060|M.marinum_M MVKGILALIADLGAD-GMTMVVVTHEMGFARSTSDAVVFMDHGTVVEAGP
MAV_0058|M.avium_104 MVKGILQLIADLGAE-GMTMVVVTHEMGFARSASDAVIFMDHGKVVESGP
Mflv_3968|M.gilvum_PYR-GCK MINEVLAVMTGLAGD-GMTMLVVTHEMGFARRAANRVVFMSDGAVVEDAE
MAB_2803c|M.abscessus_ATCC_199 TTSDVLRLLREVNAEFGVTIVVITHEMDVVRTVADHVAVLADGKVVETGT
TH_2538|M.thermoresistible__bu TTASVVELLRELRDDLNLSIVFITHEMDTVLKIADSVARLDHGRIVEAGR
Mvan_3884|M.vanbaalenii_PYR-1 TTRSILALLSHLRDEFGLSIILITHEMEVVREIADSVARIEDGRIIESGS
MSMEG_3056|M.smegmatis_MC2_155 TTQEILTLLTNLRDELDVTVLLITHDLSIVRQIADRVTVLSEGRVVAQGS
MLBr_00669|M.leprae_Br4923 TSKDIMDLLERINRT-GTTVLMATHDHHIVDSMRQRVVELSLGRLVR-DE
Mb1312c|M.bovis_AF2122/97 VQAQILDVLKAARDVTGAGVLIITHDLGVVAEFADRALVMYAGRVVESAG
Rv1281c|M.tuberculosis_H37Rv VQAQILDVLKAARDVTGAGVLIITHDLGVVAEFADRALVMYAGRVVESAG
MUL_4001|M.ulcerans_Agy99 VQAQILDVLKTARDVTGAGVLIITHDLGVVAEFADRALVMYAGRVVEKAT
:: :: . ::. **: . : . : * ::
MMAR_0060|M.marinum_M PEQLFEAARTERLQRFLAQVL-----------------------------
MAV_0058|M.avium_104 PEQIFEAAETERLQRFLSQVL-----------------------------
Mflv_3968|M.gilvum_PYR-GCK PIQFFDNPQTDRAKDFLGKILHH---------------------------
MAB_2803c|M.abscessus_ATCC_199 VFDVFSTPQSEAGKRFVGTVLRNTPSGEDIARIAQRHKG-----------
TH_2538|M.thermoresistible__bu LVDLLTDPASALGADLRPQGAAAPPAPPX-XXRLRGVAG-----------
Mvan_3884|M.vanbaalenii_PYR-1 VQDVILDPASELARELLPDRPSVPLDGAG-EIWEVSHAS-----------
MSMEG_3056|M.smegmatis_MC2_155 LADVAGDPASG----LLPPLSDPPPPTDD-ELILNVHAG-----------
MLBr_00669|M.leprae_Br4923 WCGIYGMDR-----------------------------------------
Mb1312c|M.bovis_AF2122/97 VNDLYRDRRMPYTVGLLGSVPRLDAAQGTRLVPIPGAPPSLAGLAPGCPF
Rv1281c|M.tuberculosis_H37Rv VNDLYRDRRMPYTVGLLGSVPRLDAAQGTRLVPIPGAPPSLAGLAPGCPF
MUL_4001|M.ulcerans_Agy99 VNDLYHNRRMPYTIGLLGSVPRLDATQGTRLVPIPGAPPSPAGLDRGCPF
.
MMAR_0060|M.marinum_M --------------------------------------------------
MAV_0058|M.avium_104 --------------------------------------------------
Mflv_3968|M.gilvum_PYR-GCK --------------------------------------------------
MAB_2803c|M.abscessus_ATCC_199 -RIVAVEISEGVQIGPALARATELGVRFEVVYGGITTLQSRSFGNLTLEL
TH_2538|M.thermoresistible__bu -PDHPHR--TGAVRAAGRTGPAAARRCGGGTAVD-AGSDRRVADRAGRRP
Mvan_3884|M.vanbaalenii_PYR-1 -RTVPLDWLTSIQSAPGLSGAAVSVLSASVESIRGVAVGRAVLAITPSAP
MSMEG_3056|M.smegmatis_MC2_155 -ADASGAFIGALVRELN---TDVRILDGEVLRLGGQSVSQFQLALTGIDH
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
Mb1312c|M.bovis_AF2122/97 APRCPLVIDECLTAEPELLDVATDHRAACIRTELVTGRSAADIYRVKTEA
Rv1281c|M.tuberculosis_H37Rv APRCPLVIDECLTAEPELLDVATDHRAACIRTELVTGRSAADIYRVKTEA
MUL_4001|M.ulcerans_Agy99 APRCPLVIDECRSDEPELIEVGQNHRAACIRTDQVNGRSAADIYGVQTEA
MMAR_0060|M.marinum_M --------------------------------------------------
MAV_0058|M.avium_104 --------------------------------------------------
Mflv_3968|M.gilvum_PYR-GCK --------------------------------------------------
MAB_2803c|M.abscessus_ATCC_199 EGPDDQVAHVVSDLSAVTTVQRVAR-------------------------
TH_2538|M.thermoresistible__bu G-------------RRDDAVAAPRRRPIRP--------------------
Mvan_3884|M.vanbaalenii_PYR-1 SGFVEYLTERGLHVRPVEKVAAGSAEAAA---------------------
MSMEG_3056|M.smegmatis_MC2_155 SAADRAEEYVRWFERNGLTAVRVGERSLV---------------------
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
Mb1312c|M.bovis_AF2122/97 RPAALGDASVVVRVRHLVKTYRLAKGVVLRRAIGEVRAVDGISLELRQGR
Rv1281c|M.tuberculosis_H37Rv RPAALGDASVVVRVRHLVKTYRLAKGVVLRRAIGEVRAVDGISLELRQGR
MUL_4001|M.ulcerans_Agy99 TEPAPADSSVVVRVSDLVKTYPLTKGVLLRRSIGEVRAVDGVSFELRQGH
MMAR_0060|M.marinum_M --------------------------------------------------
MAV_0058|M.avium_104 --------------------------------------------------
Mflv_3968|M.gilvum_PYR-GCK --------------------------------------------------
MAB_2803c|M.abscessus_ATCC_199 --------------------------------------------------
TH_2538|M.thermoresistible__bu --------------------------------------------------
Mvan_3884|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3056|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
Mb1312c|M.bovis_AF2122/97 TLGIVGESGSGKSTTLHEILELAAPQSGSIEVLGTDVATLGTAERRSLRR
Rv1281c|M.tuberculosis_H37Rv TLGIVGESGSGKSTTLHEILELAAPQSGSIEVLGTDVATLGTAERRSLRR
MUL_4001|M.ulcerans_Agy99 TLGIVGESGSGKSTTLHQILELAAPQSGSIEVLGTDVTTLTASSRRALRR
MMAR_0060|M.marinum_M --------------------------------------------------
MAV_0058|M.avium_104 --------------------------------------------------
Mflv_3968|M.gilvum_PYR-GCK --------------------------------------------------
MAB_2803c|M.abscessus_ATCC_199 --------------------------------------------------
TH_2538|M.thermoresistible__bu --------------------------------------------------
Mvan_3884|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3056|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
Mb1312c|M.bovis_AF2122/97 DIQVVFQDPVASLDPRLPVFDLIAEPLQANGFGKNETHARVAELLDIVGL
Rv1281c|M.tuberculosis_H37Rv DIQVVFQDPVASLDPRLPVFDLIAEPLQANGFGKNETHARVAELLDIVGL
MUL_4001|M.ulcerans_Agy99 DIQVVFQDPVASLDPRLPISDLLAEPLQANGFNKTDTQDRVAELLDIVGL
MMAR_0060|M.marinum_M --------------------------------------------------
MAV_0058|M.avium_104 --------------------------------------------------
Mflv_3968|M.gilvum_PYR-GCK --------------------------------------------------
MAB_2803c|M.abscessus_ATCC_199 --------------------------------------------------
TH_2538|M.thermoresistible__bu --------------------------------------------------
Mvan_3884|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3056|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
Mb1312c|M.bovis_AF2122/97 RHGDASRYPAEFSGGQKQRIGIARALALQPKILALDEPVSALDVSIQAGI
Rv1281c|M.tuberculosis_H37Rv RHGDASRYPAEFSGGQKQRIGIARALALQPKILALDEPVSALDVSIQAGI
MUL_4001|M.ulcerans_Agy99 RRSDASRYPAEFSGGQKQRIGIARALALQPKILALDEPVSALDVSIQAGI
MMAR_0060|M.marinum_M --------------------------------------------------
MAV_0058|M.avium_104 --------------------------------------------------
Mflv_3968|M.gilvum_PYR-GCK --------------------------------------------------
MAB_2803c|M.abscessus_ATCC_199 --------------------------------------------------
TH_2538|M.thermoresistible__bu --------------------------------------------------
Mvan_3884|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3056|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
Mb1312c|M.bovis_AF2122/97 INLLLDLQEQFGLSYLFVSHDLSVVKHLAHQVAVMLAGTVVEQGDSEEVF
Rv1281c|M.tuberculosis_H37Rv INLLLDLQEQFGLSYLFVSHDLSVVKHLAHQVAVMLAGTVVEQGDSEEVF
MUL_4001|M.ulcerans_Agy99 INLLLDLQAGFKLSYLFVSHDLSVVKHLANSVVVMFAGSIVEQGNSQEVF
MMAR_0060|M.marinum_M ------------------------
MAV_0058|M.avium_104 ------------------------
Mflv_3968|M.gilvum_PYR-GCK ------------------------
MAB_2803c|M.abscessus_ATCC_199 ------------------------
TH_2538|M.thermoresistible__bu ------------------------
Mvan_3884|M.vanbaalenii_PYR-1 ------------------------
MSMEG_3056|M.smegmatis_MC2_155 ------------------------
MLBr_00669|M.leprae_Br4923 ------------------------
Mb1312c|M.bovis_AF2122/97 GNPKHEYTRRLLGAVPQPDPARRG
Rv1281c|M.tuberculosis_H37Rv GNPKHEYTRRLLGAVPQPDPARRG
MUL_4001|M.ulcerans_Agy99 SNPQHEYTQRLLGAVPQPIPGNAD