For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VMSIYTEDLTKFYGTTRALEGVSISVDQGEILGVVGSSGSGKSTLVRNIALLERPTRGRVVLDGQDLTAL SEKELRGARRRLGNVFQSANLLDNRTARANIEYPLEIAGWDRKKRWYRAQELLELVGLPGRGDAYPAQLS GGQQQRVGIARALAAHPTVILADEPTSALDPTTTQEILTLLTNLRDELDVTVLLITHDLSIVRQIADRVT VLSEGRVVAQGSLADVAGDPASGLLPPLSDPPPPTDDELILNVHAGADASGAFIGALVRELNTDVRILDG EVLRLGGQSVSQFQLALTGIDHSAADRAEEYVRWFERNGLTAVRVGERSLV
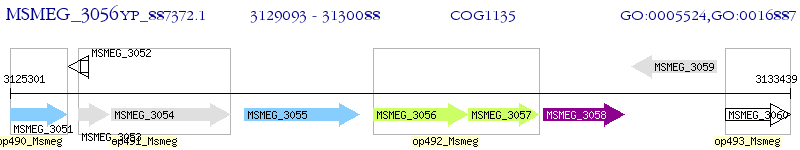
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3056 | - | - | 100% (331) | ABC transporter ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_5807 | - | 1e-57 | 51.67% (240) | D-methionine transport ATP-binding protein MetN |
| M. smegmatis MC2 155 | MSMEG_6120 | - | 1e-45 | 43.75% (240) | peptide transport system ATP-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2419c | cysA1 | 1e-41 | 40.34% (233) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. gilvum PYR-GCK | Mflv_3581 | - | 2e-44 | 40.93% (237) | ABC transporter related |
| M. tuberculosis H37Rv | Rv2397c | cysA1 | 1e-41 | 40.34% (233) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. leprae Br4923 | MLBr_01122 | - | 2e-39 | 41.15% (226) | putative ABC transport protein, ATP-binding component |
| M. abscessus ATCC 19977 | MAB_2803c | - | 7e-63 | 45.07% (304) | methionine import ATP-binding protein metN |
| M. marinum M | MMAR_3715 | cysA1 | 7e-41 | 39.91% (233) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. avium 104 | MAV_1785 | - | 4e-40 | 39.32% (234) | sulfate/thiosulfate import ATP-binding protein CysA |
| M. thermoresistible (build 8) | TH_2538 | - | 1e-60 | 51.64% (244) | PUTATIVE ABC transporter related |
| M. ulcerans Agy99 | MUL_3658 | cysA1 | 1e-40 | 39.48% (233) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. vanbaalenii PYR-1 | Mvan_3884 | - | 2e-60 | 51.67% (240) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
TH_2538|M.thermoresistible__bu -----------------MIELTDLTKVYGCGEHRTVVLDRINFRVETGEV
Mvan_3884|M.vanbaalenii_PYR-1 --MLRVTAGPSTVESFPVIEIENLTKRFG----DRTVLDDISLSVGSGEI
MSMEG_3056|M.smegmatis_MC2_155 ---------------MMSIYTEDLTKFYG----TTRALEGVSISVDQGEI
MAB_2803c|M.abscessus_ATCC_199 -----------MGDVAPLIQFQNVTKTFRVGKKTVTAVDDVSLDIGSGDV
Mb2419c|M.bovis_AF2122/97 ------MT--------YAIVVADATKRYG----DFVALDHVDFVVPTGSL
Rv2397c|M.tuberculosis_H37Rv ------MT--------YAIVVADATKRYG----DFVALDHVDFVVPTGSL
MMAR_3715|M.marinum_M MKKETSLTGPGT--GGHAIVVRDAFKQYG----DFVALDHVDFVVPAGSL
MUL_3658|M.ulcerans_Agy99 ------MTGPGT--GGHAIVVRDAFKQYG----DFVALDHVDFVVPAGSL
MAV_1785|M.avium_104 ------MTDAGTDRGDIAITVRDAYKRYG----DFVALDHVDFVVPTGSL
Mflv_3581|M.gilvum_PYR-GCK ------MTTSENS-AHPMVKAELVCKDFG----ALKVLKGITLEVQKGQV
MLBr_01122|M.leprae_Br4923 --------------MSPLLEVTDLAVTFETNGEPVTAVRGISYRINAGEV
: : .: : : *.:
TH_2538|M.thermoresistible__bu LAVVGPSGAGKSTLAQCINLLT---RPTSGSVVVNGE---------DLTT
Mvan_3884|M.vanbaalenii_PYR-1 LAVVGPSGAGKSTLSRCVSFLE---RPTSGTVRVDGK---------DFTR
MSMEG_3056|M.smegmatis_MC2_155 LGVVGSSGSGKSTLVRNIALLE---RPTRGRVVLDGQ---------DLTA
MAB_2803c|M.abscessus_ATCC_199 FAMIGYSGAGKSTLVRLINGLE---RPTSGRVIVDGA---------EVSA
Mb2419c|M.bovis_AF2122/97 TALLGPSGSGKSTLLRTIAGLD---QPDTGTITING--------------
Rv2397c|M.tuberculosis_H37Rv TALLGPSGSGKSTLLRTIAGLD---QPDTGTITING--------------
MMAR_3715|M.marinum_M TALLGPSGSGKSTLLRTIAGLD---QPDSGNITING--------------
MUL_3658|M.ulcerans_Agy99 TALLGPSGSGKSTLLRTIAGLD---QPDSGNITING--------------
MAV_1785|M.avium_104 TALLGPSGSGKSTLLRTIAGLD---QPDTGTVTIYG--------------
Mflv_3581|M.gilvum_PYR-GCK LVLVGPSGSGKSTLLRCINHLE---TVSAGRLYVDGTLVGYHERGGKLYE
MLBr_01122|M.leprae_Br4923 VAMVGESGSGKSAAAMAVVGLLPEYAQVRGSVRLHNT---------ELVG
::* **:***: : * * : : .
TH_2538|M.thermoresistible__bu LSTHRLRVAR-RRIGTVFQS--SGLLERRTAAENVALP--LEYLGVTAAE
Mvan_3884|M.vanbaalenii_PYR-1 LDGDELLAAR-RNVGVIFQT--APLLRRRTVAQNIALP--LQYLHATEGS
MSMEG_3056|M.smegmatis_MC2_155 LSEKELRGAR-RRLGNVFQS--ANLLDNRTARANIEYP--LEIAGWDRKK
MAB_2803c|M.abscessus_ATCC_199 LGERELRGLR-SDIGMIFQQ--FNLFRSRTVAGNIAYP--LRVAGWTADK
Mb2419c|M.bovis_AF2122/97 RDVTRVPPQR-RGIGFVFQH--YAAFKHLTVRDNVAFG--LKIRKRPKAE
Rv2397c|M.tuberculosis_H37Rv RDVTRVPPQR-RGIGFVFQH--YAAFKHLTVRDNVAFG--LKIRKRPKAE
MMAR_3715|M.marinum_M RDVTRVPPQR-RGIGFVFQH--YAAFKHLTVRDNVAYG--LKIRKRPKAE
MUL_3658|M.ulcerans_Agy99 RDVTRVPPQR-RGIGFVFQH--YAAFKHLTVRDNVAYG--LKIRKRPKAE
MAV_1785|M.avium_104 RDVTRVPPQR-RGIGFVFQH--YAAFKHLTVRDNVAYG--LKVRKRPKAE
Mflv_3581|M.gilvum_PYR-GCK MKPRDVARQR-RDVGMVFQH--FNLFPHRTALGNVVEAP-IQVKGVKKDE
MLBr_01122|M.leprae_Br4923 LADKTMSRFRGKAIGTVFQDPMSALTPVYTVGDQITEAIKVHQPRVGKKA
: * :* :** *. :: :.
TH_2538|M.thermoresistible__bu TRKRVAELLDRVGLAA---RANHYPFQLSGGQRQRVGIARALALRPSVLL
Mvan_3884|M.vanbaalenii_PYR-1 AGTRVTELLDRVGLAD---RRDFYPAQLSGGQKQRVGIARALALGPSNLL
MSMEG_3056|M.smegmatis_MC2_155 RWYRAQELLELVGLPG---RGDAYPAQLSGGQQQRVGIARALAAHPTVIL
MAB_2803c|M.abscessus_ATCC_199 RNARVAELLEFVGLGD---KAKSYPDQLSGGQKQRVGIARALATSPSVLL
Mb2419c|M.bovis_AF2122/97 IKAKVDNLLQVVGLSG---FQSRYPNQLSGGQRQRMALARALAVDPEVLL
Rv2397c|M.tuberculosis_H37Rv IKAKVDNLLQVVGLSG---FQSRYPNQLSGGQRQRMALARALAVDPEVLL
MMAR_3715|M.marinum_M IRDKVDNLLEVVGLSG---FHGRYPNQLSGGQRQRMALARALAVDPEVLL
MUL_3658|M.ulcerans_Agy99 IRDKVDNLLEVVGLSG---FHGRYPDQLSGGQRQRMALARALAVDPEVLL
MAV_1785|M.avium_104 IKAKVDNLLEVVGLSG---FQGRYPNQLSGGQRQRMALARALAVDPQVLL
Mflv_3581|M.gilvum_PYR-GCK AVTRGRELLAQVGLAD---KADAYPAQLSGGQQQRVAIARALAMDPKLML
MLBr_01122|M.leprae_Br4923 AHRRAVELLELVGITQPDRRARAFPHELSGGERQRVVIATAIANDPDLLI
: :** **: :* :****::**: :* *:* * ::
TH_2538|M.thermoresistible__bu SDEATAGLDPTTTASVVELLRELRDDLNLSIVFITHEMDTVLKIADSVAR
Mvan_3884|M.vanbaalenii_PYR-1 SDEATSGLDPATTRSILALLSHLRDEFGLSIILITHEMEVVREIADSVAR
MSMEG_3056|M.smegmatis_MC2_155 ADEPTSALDPTTTQEILTLLTNLRDELDVTVLLITHDLSIVRQIADRVTV
MAB_2803c|M.abscessus_ATCC_199 ADEATSALDPETTSDVLRLLREVNAEFGVTIVVITHEMDVVRTVADHVAV
Mb2419c|M.bovis_AF2122/97 LDEPFGALDAKVREELRAWLRRLHDEVHVTTVLVTHDQAEALDVADRIAV
Rv2397c|M.tuberculosis_H37Rv LDEPFGALDAKVREELRAWLRRLHDEVHVTTVLVTHDQAEALDVADRIAV
MMAR_3715|M.marinum_M LDEPFGALDAKVREDLRAWLRRLHDEVHVTTVLVTHDQAEALDVADRIAV
MUL_3658|M.ulcerans_Agy99 LDEPFGALDAKVREDLRAWLRRLHDEVHVTTVLVTHDQAEALDVADRIAV
MAV_1785|M.avium_104 LDEPFGALDAKVREDLRAWLRRLHDEVHVTTVLVTHDQAEALDVADRIAV
Mflv_3581|M.gilvum_PYR-GCK FDEPTSALDPELVGEVLAVMKKLAAQG-MTMVVVTHEMGFAREVADELVF
MLBr_01122|M.leprae_Br4923 CDEPTTALDVTVQAQILEVLKTARDVTGAGVLIITHDLGVVTEFADQALV
**. .** .: : :.:**: . .**
TH_2538|M.thermoresistible__bu LDHGRIVEAGRLVDLLTDPASALGADLRPQGAAAPPAPPXXXRLRG----
Mvan_3884|M.vanbaalenii_PYR-1 IEDGRIIESGSVQDVILDPASELARELLPDRPSVPLDGAGEIWEVS----
MSMEG_3056|M.smegmatis_MC2_155 LSEGRVVAQGSLADVAGDPASG----LLPPLSDPPPPTDDELILNV----
MAB_2803c|M.abscessus_ATCC_199 LADGKVVETGTVFDVFSTPQSEAGKRFVGTVLRNTPSGEDIARIAQR---
Mb2419c|M.bovis_AF2122/97 LHKGRIEQVGSPTDVYDAPANAFVMSFLGAVSTLNGSLVRPHDIRVGRTP
Rv2397c|M.tuberculosis_H37Rv LHKGRIEQVGSPTDVYDAPANAFVMSFLGAVSTLNGSLVRPHDIRVGRTP
MMAR_3715|M.marinum_M LNSGRIEQVGSPTEVYDAPANAFVMSFLGAVSALNGALVRPHDIRVGRTP
MUL_3658|M.ulcerans_Agy99 LNSGRIEQVGSPTEVYDAPTNAFVMSFLGAVSALNGALVRPHDIRVGRTP
MAV_1785|M.avium_104 LNQGRIEQIGSPTEVYDAPTNAFVMSFLGAVSTLNGTLVRPHDIRVGRTP
Mflv_3581|M.gilvum_PYR-GCK MDEGVVVERGNPREVMSNPQHERTKAFLSKVM------------------
MLBr_01122|M.leprae_Br4923 MYAGRVVESAQVSTLYRERQMPYTVGLLGSVPRLDATQGTRLVPIPGAPP
: * : . : :
TH_2538|M.thermoresistible__bu ---------VAGPDHPHR--TGAVRAAGRT-----GPAAARRCGGGTAVD
Mvan_3884|M.vanbaalenii_PYR-1 ---------HASRTVPLDWLTSIQSAPGLS-----GAAVSVLSASVESIR
MSMEG_3056|M.smegmatis_MC2_155 ---------HAGADASGAFIGALVRELN--------TDVRILDGEVLRLG
MAB_2803c|M.abscessus_ATCC_199 ---------HKGRIVAVEISEGVQIGPALARATELGVRFEVVYGGITTLQ
Mb2419c|M.bovis_AF2122/97 NMA------VAAADGTAGSTGVLRAVVDRVVVLGFEVRVELTSAATGGAF
Rv2397c|M.tuberculosis_H37Rv NMA------VAAADGTAGSTGVLRAVVDRVVVLGFEVRVELTSAATGGAF
MMAR_3715|M.marinum_M EMA------VAAADGTGESTGVTRAVVDRIVVLGFEVRVELTSAATGGAF
MUL_3658|M.ulcerans_Agy99 EMA------VAAADGTGESTGVTRAVVDRIVVLGFEVRVELTSAATGGAF
MAV_1785|M.avium_104 EMA------VAAEDGTAESTGVARAIVDRVVKLGFEVRVELTSAATGGPF
Mflv_3581|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_01122|M.leprae_Br4923 SLAGLQQECPFAPRCPLVVDECRLREPELVEVRAGHRTACIRTDQVSGRS
TH_2538|M.thermoresistible__bu -AGSDRRVADRAGRRPG-------------RRDDAVAAPRRRPIRP----
Mvan_3884|M.vanbaalenii_PYR-1 GVAVGRAVLAITPSAPSGFVEYLTERGLHVRPVEKVAAGSAEAAA-----
MSMEG_3056|M.smegmatis_MC2_155 GQSVSQFQLALTGIDHSAADRAEEYVRWFERNGLTAVRVGERSLV-----
MAB_2803c|M.abscessus_ATCC_199 SRSFGNLTLELEGPDDQ---------VAHVVSDLSAVTTVQRVAR-----
Mb2419c|M.bovis_AF2122/97 TAQITRGDAEALALREGDTVYVRATRVPPIAG---GVSGVDDAGVERVKV
Rv2397c|M.tuberculosis_H37Rv TAQITRGDAEALALREGDTVYVRATRVPPIAG---GVSGVDDAGVERVKV
MMAR_3715|M.marinum_M TAQITRGDAEALALQEGDTVYVRATRVPPIAG---AAPAVPDDGAGSTPS
MUL_3658|M.ulcerans_Agy99 TAQITRGDAEALALQEGDTVYVRATRVPPIAG---AAPAVPDDGAGSTPR
MAV_1785|M.avium_104 TAQITRGDAEALALREGDTVYVRATRVPPITAGATTVPAVSRDGADEATL
Mflv_3581|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_01122|M.leprae_Br4923 AADIYGINTDTRTSATSDSSVVVRVRDLVKTYRLTKGVGLRRAIGEIRAV
TH_2538|M.thermoresistible__bu --------------------------------------------------
Mvan_3884|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3056|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2803c|M.abscessus_ATCC_199 --------------------------------------------------
Mb2419c|M.bovis_AF2122/97 TST-----------------------------------------------
Rv2397c|M.tuberculosis_H37Rv TST-----------------------------------------------
MMAR_3715|M.marinum_M AAQIGVGGQ-----------------------------------------
MUL_3658|M.ulcerans_Agy99 AAQIGVGGQ-----------------------------------------
MAV_1785|M.avium_104 TSA-----------------------------------------------
Mflv_3581|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_01122|M.leprae_Br4923 NGVSFELQQGRTLGIVGESGSGKSTTLHEILRLAVPQSGSIEVLGIDVAT
TH_2538|M.thermoresistible__bu --------------------------------------------------
Mvan_3884|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3056|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2803c|M.abscessus_ATCC_199 --------------------------------------------------
Mb2419c|M.bovis_AF2122/97 --------------------------------------------------
Rv2397c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3715|M.marinum_M --------------------------------------------------
MUL_3658|M.ulcerans_Agy99 --------------------------------------------------
MAV_1785|M.avium_104 --------------------------------------------------
Mflv_3581|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_01122|M.leprae_Br4923 LNRASRRSMRRNIQVVFQDPVASLDPRLPVFELIAEPLHANGFSKNYTQM
TH_2538|M.thermoresistible__bu --------------------------------------------------
Mvan_3884|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3056|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2803c|M.abscessus_ATCC_199 --------------------------------------------------
Mb2419c|M.bovis_AF2122/97 --------------------------------------------------
Rv2397c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3715|M.marinum_M --------------------------------------------------
MUL_3658|M.ulcerans_Agy99 --------------------------------------------------
MAV_1785|M.avium_104 --------------------------------------------------
Mflv_3581|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_01122|M.leprae_Br4923 RVAELLDIVGLRRDDATRYPTEFSGGQKQRIGIARALALQPKILAFDEPV
TH_2538|M.thermoresistible__bu --------------------------------------------------
Mvan_3884|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3056|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2803c|M.abscessus_ATCC_199 --------------------------------------------------
Mb2419c|M.bovis_AF2122/97 --------------------------------------------------
Rv2397c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3715|M.marinum_M --------------------------------------------------
MUL_3658|M.ulcerans_Agy99 --------------------------------------------------
MAV_1785|M.avium_104 --------------------------------------------------
Mflv_3581|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_01122|M.leprae_Br4923 SALDVSIQAGIINLLLDLQDRLGLSYLFVSHDLSVIKHLAHQVAVMLAGT
TH_2538|M.thermoresistible__bu --------------------------------
Mvan_3884|M.vanbaalenii_PYR-1 --------------------------------
MSMEG_3056|M.smegmatis_MC2_155 --------------------------------
MAB_2803c|M.abscessus_ATCC_199 --------------------------------
Mb2419c|M.bovis_AF2122/97 --------------------------------
Rv2397c|M.tuberculosis_H37Rv --------------------------------
MMAR_3715|M.marinum_M --------------------------------
MUL_3658|M.ulcerans_Agy99 --------------------------------
MAV_1785|M.avium_104 --------------------------------
Mflv_3581|M.gilvum_PYR-GCK --------------------------------
MLBr_01122|M.leprae_Br4923 IVEQGDSHDVFRNPQHDYTRRLLGAIPRPEVG