For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPVHTAQQAVSNHANPKPTSQVQGGLLDPKMLWRSLPDALRKLNPRTLWRNPVMFIVEVGAVWSTILAGV NPTWFGWLIVAWLWLTVVFANLAEAVAEGRGKAQAESLRKTKADTMARRLTESGAEEAVPAPQLRQGDIV VVEAGQTIPGDGDVVEGIASVDESAITGESAPVIRESGGDRSAVTGGTTVLSDRIVVRITQKPGESFIDR MIALVEGANRQKTPNEIALNILLAALTIIFVFAVATLQPLAIYAKANNPGVPDSLALTGNGISGIVMVAL LVCLIPTTIGALLSAIGIAGMDRLVQRNVLAMSGRAVEAAGDVNTLLLDKTGTITLGNRQAAAFVPLTGV TPESLADAAQLSSLADETPEGRSIVVFAKREFGLRARTPGELAEAHWIEFSATTRMSGVNLGEHRLRKGA AAAVVDWVRGEGGTVPVELFDVVDAISHAGGTPLVVGHVITDDTDPKRAEVLGVIHLKDVVKQGMKQRFD EMRRMGIRTVMITGDNPLTAKAIADEAGVDDFLAEATPEDKLALIKQEQAGGRLVAMTGDGTNDAPALAQ ADVGVAMNTGTSAAKEAGNMVDLDSDPTKLIEIVEIGKQLLITRGALTTFSISNDIAKYFAIIPALFVGL FPGLDLLNIMRLHSPQSAILSAVIFNAIVIVALIPLALKGVRYTPSSAAKLLSRNLYIYGLGGIIAPFLG IKLIDLVVQLLPGMS
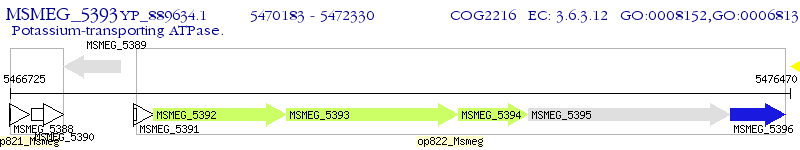
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5393 | kdpB | - | 100% (715) | potassium-transporting ATPase subunit B |
| M. smegmatis MC2 155 | MSMEG_5014 | - | 3e-56 | 31.77% (513) | copper-translocating P-type ATPase |
| M. smegmatis MC2 155 | MSMEG_6058 | - | 1e-41 | 30.06% (519) | cadmium transporting P-type ATPase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1059 | kdpB | 0.0 | 70.59% (697) | potassium-transporting ATPase subunit B |
| M. gilvum PYR-GCK | Mflv_2849 | - | 3e-51 | 31.07% (515) | heavy metal translocating P-type ATPase |
| M. tuberculosis H37Rv | Rv1030 | kdpB | 0.0 | 70.59% (697) | potassium-transporting ATPase subunit B |
| M. leprae Br4923 | MLBr_02000 | ctpB | 7e-46 | 29.55% (494) | cation-transporting ATPase |
| M. abscessus ATCC 19977 | MAB_3253c | - | 0.0 | 67.28% (703) | potassium-transporting ATPase B chain |
| M. marinum M | MMAR_0632 | kdpB | 0.0 | 85.37% (697) | high-affinity K+ transport system, ATPase chain B, KdpB |
| M. avium 104 | MAV_1172 | kdpB | 0.0 | 83.87% (713) | potassium-transporting ATPase subunit B |
| M. thermoresistible (build 8) | TH_4361 | - | 3e-50 | 29.95% (561) | PUTATIVE heavy metal translocating P-type ATPase |
| M. ulcerans Agy99 | MUL_0423 | ctpV | 3e-52 | 29.38% (582) | metal cation transporter p-type ATPase, CtpV |
| M. vanbaalenii PYR-1 | Mvan_4760 | - | 0.0 | 85.67% (719) | potassium-transporting ATPase subunit B |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1059|M.bovis_AF2122/97 --------------------------------------------------
Rv1030|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_5393|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_4760|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0632|M.marinum_M --------------------------------------------------
MAV_1172|M.avium_104 --------------------------------------------------
MAB_3253c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2849|M.gilvum_PYR-GCK ---------MTTSIPVSGPSIELRIGGMTCASCANRIERKLNKID----G
TH_4361|M.thermoresistible__bu ---------MNTGG---MNTVTLTVGGMTCASCAARVEKRLNRID----G
MLBr_02000|M.leprae_Br4923 --MTASLVEDTNNNHESVRRIQLDVAGMLCAACASRVETKLNKIP----G
MUL_0423|M.ulcerans_Agy99 MRVHANGFAIDATRAAAIEESVGRVGGVRAVRAYPRTASVVIWYSPAHCD
Mb1059|M.bovis_AF2122/97 -----MMIARMETSATAAAATSAPRLRLAKRS------------------
Rv1030|M.tuberculosis_H37Rv -----MMIARMETSATAAAATSAPRLRLAKRS------------------
MSMEG_5393|M.smegmatis_MC2_155 ------MPVHTAQQAVSNHANPKP-TSQVQGG------------------
Mvan_4760|M.vanbaalenii_PYR-1 ------MSVTTAGAAARKHAEPKS-SNKIQGG------------------
MMAR_0632|M.marinum_M -----------------------M-STTVGSG------------------
MAV_1172|M.avium_104 ------MTVTAIDPAEQAHPAPSAGTKRVQGG------------------
MAB_3253c|M.abscessus_ATCC_199 --------------------MSKSKNAVVTTG------------------
Mflv_2849|M.gilvum_PYR-GCK VAATVNYATEKATVTVPDGYDPALLIAEVQNAGYSAALPTPEK-------
TH_4361|M.thermoresistible__bu VVATVNFATEQATVRYPDHVDPADLIAAIEATGYTASLPAPQRDTGGAPA
MLBr_02000|M.leprae_Br4923 VRASVNFATRVATIDAVD-VAVDELRQVIEQAGYRATA------------
MUL_0423|M.ulcerans_Agy99 TAAIVSAIAEAEHAGDASAPAPAARVAGIADGGVVGRVIGGLAR---ALL
Mb1059|M.bovis_AF2122/97 --------------------------------------------------
Rv1030|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_5393|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_4760|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0632|M.marinum_M --------------------------------------------------
MAV_1172|M.avium_104 --------------------------------------------------
MAB_3253c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2849|M.gilvum_PYR-GCK PALTGETGETEDPDLVS---------------LRHRLTGS----------
TH_4361|M.thermoresistible__bu GAATGAAGDDHDAEAER---------------WRRRLTIS----------
MLBr_02000|M.leprae_Br4923 HAESAVEEIDPDADYARN--------------LLRRLIVA----------
MUL_0423|M.ulcerans_Agy99 GMRTDDNDDDPPPEIARLRAALDRIGYSPEQDTATQLSPSEQARSGIRFW
Mb1059|M.bovis_AF2122/97 ----------------------------------LFDPMIVRSALPQSLR
Rv1030|M.tuberculosis_H37Rv ----------------------------------LFDPMIVRSALPQSLR
MSMEG_5393|M.smegmatis_MC2_155 ----------------------------------LLDPKMLWRSLPDALR
Mvan_4760|M.vanbaalenii_PYR-1 ----------------------------------LLDPNMLWKSLPSALR
MMAR_0632|M.marinum_M ----------------------------------LLDPTMLWKSLPAAVR
MAV_1172|M.avium_104 ----------------------------------LLDPKMLWRSTPDALR
MAB_3253c|M.abscessus_ATCC_199 ----------------------------------VFDPAQLIKALPLAVR
Mflv_2849|M.gilvum_PYR-GCK ---VLLSVPVIAMA----MIPVLQFTYWQWASLVLAAPVIVWAAWPFHRA
TH_4361|M.thermoresistible__bu ---AALSIPVVVLS----MIPALQFTHWQWLALTLTSPVVVWGAWPFHRA
MLBr_02000|M.leprae_Br4923 ---ALLFVPLADLSTMFAIVPTNRFPGWGYLLTALAAPIVTWAAWPFHRV
MUL_0423|M.ulcerans_Agy99 SRRAWLAWPLGLLASG-LTMFFGAYPWAGWLAFAATIPVQFIAGWPFLVG
* . *
Mb1059|M.bovis_AF2122/97 KLAPRVQARNPVMLVVLVGAVITTLAFLRDLASSTAQ-------------
Rv1030|M.tuberculosis_H37Rv KLAPRVQARNPVMLVVLVGAVITTLAFLRDLASSTAQ-------------
MSMEG_5393|M.smegmatis_MC2_155 KLNPRTLWRNPVMFIVEVGAVWSTILAGVNPT------------------
Mvan_4760|M.vanbaalenii_PYR-1 KLDPRTMWRNPVMFIVEIGAVWSTILAFLEPS------------------
MMAR_0632|M.marinum_M KLDPRTLWRNPVMFIVELGAVWSTLLAVVDPS------------------
MAV_1172|M.avium_104 KLDPRTLWRNPVMFIVEIGAAWSTVLAIVGPT------------------
MAB_3253c|M.abscessus_ATCC_199 KLDPRHMARNPVMFVVTVGSVATTVLAVLHPS------------------
Mflv_2849|M.gilvum_PYR-GCK AWANLRHATATMDTLISLGTLSAFLWSLYALFFGTAGTPGMKHPFELTLA
TH_4361|M.thermoresistible__bu AVANLRHGTATMDTLISLGVTAAYLWSLWALFFTAAGMPGMTMTFTL-LP
MLBr_02000|M.leprae_Br4923 ALRNARYRAASMETLISAGILAATGWSLSTIFVDKE--PRQTHGIWQAIL
MUL_0423|M.ulcerans_Agy99 AVQQARARSANMNTLISLGTLTAFIYSTYQLFAGGP--------------
: :: * :
Mb1059|M.bovis_AF2122/97 ----ENVFNGLVAAFLWFTVLFANFAEAMAEGRGKAQAAALRKVRSETMA
Rv1030|M.tuberculosis_H37Rv ----ENVFNGLVAAFLWFTVLFANFAEAMAEGRGKAQAAALRKVRSETMA
MSMEG_5393|M.smegmatis_MC2_155 ------WFGWLIVAWLWLTVVFANLAEAVAEGRGKAQAESLRKTKADTMA
Mvan_4760|M.vanbaalenii_PYR-1 ------WFAWLIVVWLWLTVLFANLAEAVAEGRGKAQAESLRKTKTATPA
MMAR_0632|M.marinum_M ------WFAGLIVAWLWLTVIFANLAEAVAEGRGKAQADTLRKTKADTMA
MAV_1172|M.avium_104 ------WFAWLTVIWLWLTVLFANLAEAVAEGRGKAQAETLRRAKTQTMA
MAB_3253c|M.abscessus_ATCC_199 ------IFAWAITAWLWFTVVFANLAEAVAEGRGKAQAASLREVKRDTVA
Mflv_2849|M.gilvum_PYR-GCK PSHGAANIYLEVAAGVTTFILAGRYFEKRSKRQAGAALRALLEMGAKEVS
TH_4361|M.thermoresistible__bu ADTGEPHIYLEVAAAVTTFIIAGRYFEARAKRQSGAALRALLDMGANDVA
MLBr_02000|M.leprae_Br4923 HSD---SIYFEVAAGVTVFVLAGRFFEARAKSKAGSALRALAARGAKNVE
MUL_0423|M.ulcerans_Agy99 -------LFFDTAALIIAFVVLGRYFEATAHGKTREAISKLLDMGAKETR
: . : :: .. * :. : *
Mb1059|M.bovis_AF2122/97 NRRT-----AAGNIESVPSSRLDLDDVVEVSAGETIPSDGEIIEGIASVD
Rv1030|M.tuberculosis_H37Rv NRRT-----AAGNIESVPSSRLDLDDVVEVSAGETIPSDGEIIEGIASVD
MSMEG_5393|M.smegmatis_MC2_155 RRLT-----ESGAEEAVPAPQLRQGDIVVVEAGQTIPGDGDVVEGIASVD
Mvan_4760|M.vanbaalenii_PYR-1 RRLTGWAPGTPGREEAVAAALLQPGDIVVVEAGQVIPGDGDVVEGIASVD
MMAR_0632|M.marinum_M RLLT-----ATGAEEQVTAALLQPGDIVVVDAGQVIPGDGDVVEGIASVD
MAV_1172|M.avium_104 RRLRDWAPGSTGIEEAVAATALQQGDIVVVEAGQVIPGDGDVVEGIASVD
MAB_3253c|M.abscessus_ATCC_199 RRLVG-DGNGNENEEEVPGSALRPGDRVVVEAGQVIPGDGDVVEGIATVD
Mflv_2849|M.gilvum_PYR-GCK VLR-------DGVESKIAVEDLVVGDEFVVRPGEKIATDGVVASGTSAVD
TH_4361|M.thermoresistible__bu VLR-------DGAEKRVPIGELHVGDVFVVRPGEKIATDGEVVAGNSAVD
MLBr_02000|M.leprae_Br4923 VLLP------NGAELTIPAGELKKQQHFLVRPGETITADGVVIDGTATID
MUL_0423|M.ulcerans_Agy99 LIV-------DGEEHLVPVDRVQVGDLVRVRPGEKIPVDGEVIDGRAAVD
:. : : . * .*: *. ** : * :::*
Mb1059|M.bovis_AF2122/97 ESAITGESAPVIRESGGDRSAVTGGTVVLSDRIVVRITAKQGQTFIDRMI
Rv1030|M.tuberculosis_H37Rv ESAITGESAPVIRESGGDRSAVTGGTVVLSDRIVVRITAKQGQTFIDRMI
MSMEG_5393|M.smegmatis_MC2_155 ESAITGESAPVIRESGGDRSAVTGGTTVLSDRIVVRITQKPGESFIDRMI
Mvan_4760|M.vanbaalenii_PYR-1 ESAITGESAPVIRESGGDRSAVTGGTTVLSDRIVVKITQKPGESFIDRMI
MMAR_0632|M.marinum_M ESAITGESAPVIRESGGDRSAVTGGTTVLSDRILVKITQKPGESFIDKMI
MAV_1172|M.avium_104 ESAITGESAPVIRESGGDRSAVTGGTTVLSDRIVVQITQKPGESFIDRMI
MAB_3253c|M.abscessus_ATCC_199 ESAITGESAPVVRESGGDRCAVTGGTTVLSDRIVVQITAAPGESFVDRMI
Mflv_2849|M.gilvum_PYR-GCK ASMLTGESVPVEVGPG---DTVVGATVNAGGRLVVRAARVGSDTQLAQMA
TH_4361|M.thermoresistible__bu ASMLTGESVPVEVGPG---DQVVGATVNVGGHLQVRATRVGADTQLAQMA
MLBr_02000|M.leprae_Br4923 MSAITGEARPVHASPA---STVVGGTTVLDGRLVIEATAVGGDTQFAAMV
MUL_0423|M.ulcerans_Agy99 ESMLTGESVPVEKGVG---DWVAGATVNTDGLLTVRATAVGADTALAQIV
* :***: ** . *.*.*. .. : :. : .:: . :
Mb1059|M.bovis_AF2122/97 ALVEGAA-RQQTPNEIALNILLAGLTIIFLLAVVTLQPFAIYS-------
Rv1030|M.tuberculosis_H37Rv ALVEGAA-RQQTPNEIALNILLAGLTIIFLLAVVTLQPFAIYS-------
MSMEG_5393|M.smegmatis_MC2_155 ALVEGAN-RQKTPNEIALNILLAALTIIFVFAVATLQPLAIYAKANNPGV
Mvan_4760|M.vanbaalenii_PYR-1 ALVEGAN-RQKTPNEIALNILLASLTIIFVFAVTTLQPLAIYSKMNNPGV
MMAR_0632|M.marinum_M ALVEGAD-RQKTPNEIALNILLASLTIIFVFTVVTLQPLAIYSKANNPGV
MAV_1172|M.avium_104 ALVEGAN-RQKTPNEIALNILLAALTIIFVFAVATLQPLAIYSKVNNPGV
MAB_3253c|M.abscessus_ATCC_199 ALVEGAS-RQKTPNEIALNILLASLTIIFLLAVVALGPMGNYG-------
Mflv_2849|M.gilvum_PYR-GCK QLVENAQTGKAQVQRLADRISGVFVPIVLAIAVITLGAWLG---------
TH_4361|M.thermoresistible__bu RLVTEAQSGKAEVQRLADRVSAVFVPIVIALSALTLAGWLLL--------
MLBr_02000|M.leprae_Br4923 RLVEDAQVQKARVQHLADRIAAVFVPMVFVIAGLAGASWLL---------
MUL_0423|M.ulcerans_Agy99 RLVEEAQGGKAPVQRLADRVSAVFVPVVVRIAAATFAGWTLL--------
** * : :.:* .: . :.::. :: :
Mb1059|M.bovis_AF2122/97 -------GGGQRVVVLVALLVCLIPTTIGALLSAIGIAGMDRLVQHNVLA
Rv1030|M.tuberculosis_H37Rv -------GGGQRVVVLVALLVCLIPTTIGALLSAIGIAGMDRLVQHNVLA
MSMEG_5393|M.smegmatis_MC2_155 PDSLALTGNGISGIVMVALLVCLIPTTIGALLSAIGIAGMDRLVQRNVLA
Mvan_4760|M.vanbaalenii_PYR-1 PDSLALNGNGITGIVMVSLLVCLIPTTIGALLSAIGIAGMDRLVQRNVLA
MMAR_0632|M.marinum_M ADTVALNANGISGIVMVSLLVCLIPTTIGALLSAIGIAGMDRLVQRNILA
MAV_1172|M.avium_104 PDTQALNTSGVTGIVMVSLLVCLIPTTIGALLSAIGIAGMDRLVQRNVLA
MAB_3253c|M.abscessus_ATCC_199 -------GEQQDPIKLIALLVCLIPTTIGALMSAIGIAGMDRLVQHNVLA
Mflv_2849|M.gilvum_PYR-GCK ----AGFPVAAAFTAAVAVLVIACPCALGLATPTALLVGTGRGAQMGVLI
TH_4361|M.thermoresistible__bu ----TDAPVSAAFTAAVAVLIIACPCALGLATPTALLVGTGRGAQLGILI
MLBr_02000|M.leprae_Br4923 ----AGASPDRAFSVVLGVLVIACPCTLGLATPTAMMVASGRGAQLGIFI
MUL_0423|M.ulcerans_Agy99 ----AANP-VAGMTAAVSVLIIACPYALGLATPTAIIVGTGRGADMGILV
:.:*: * ::* .: :.. .* .: .::
Mb1059|M.bovis_AF2122/97 TSGRAVEAAGDVNTLLLDKTGTITLGNRQATEFVPINGVSAEAVADAAQL
Rv1030|M.tuberculosis_H37Rv TSGRAVEAAGDVNTLLLDKTGTITLGNRQATEFVPINGVSAEAVADAAQL
MSMEG_5393|M.smegmatis_MC2_155 MSGRAVEAAGDVNTLLLDKTGTITLGNRQAAAFVPLTGVTPESLADAAQL
Mvan_4760|M.vanbaalenii_PYR-1 MSGRAVEAAGDVNTLLLDKTGTITLGNRQASEFVPLTGVTPEALADAAQL
MMAR_0632|M.marinum_M MSGRAVEAAGDVNTLLLDKTGTITLGNRQAGAFVHLPGVSEAELADAAQL
MAV_1172|M.avium_104 MSGRAVEAAGDVNTLLLDKTGTITLGNRQAAAFIPLAGVPPEELADAAQL
MAB_3253c|M.abscessus_ATCC_199 KSGRAVEAAGDIDTLLMDKTGTITFGNRQATEFLVAPGIPHETLAAAARA
Mflv_2849|M.gilvum_PYR-GCK KGPEVLESTRKVDTVVLDKTGTVTTGKMTLVDVVTAPGTDRGNLLRLAGA
TH_4361|M.thermoresistible__bu KGPQVLESTRRIDTVVLDKTGTVTTGQMSVTSVHVADGLSETELLRLAGA
MLBr_02000|M.leprae_Br4923 KGYRALETINAIDTVVFDKTGTLTLGQLSVSTVTSTGGWCSGEVLALASA
MUL_0423|M.ulcerans_Agy99 KGGEVLEASKKIDTVVFDKTGTLTRAQMELTDVIAGKRRQPDVVLRIAAA
. ..:*: ::*:::*****:* .: . : *
Mb1059|M.bovis_AF2122/97 SSLADETPEGRSIVVLAKDEFGLRARDEGVMSHARFVPFTAETRMSGVDL
Rv1030|M.tuberculosis_H37Rv SSLADETPEGRSIVVLAKDEFGLRARDEGVMSHARFVPFTAETRMSGVDL
MSMEG_5393|M.smegmatis_MC2_155 SSLADETPEGRSIVVFAKREFGLRARTPGELAEAHWIEFSATTRMSGVNL
Mvan_4760|M.vanbaalenii_PYR-1 SSLADETPEGRSIVVFAKQEYGLRARTPGELAHAAWIEFSATTRMSGVDV
MMAR_0632|M.marinum_M SSLADETPEGRSIVVYAKQQYGLRARTPGELAHAQWIEFSATTRMSGVNV
MAV_1172|M.avium_104 SSLADETPEGRSVVVFAKQHFGLRARTPGELSQAQWVAFSATTRMSGVDL
MAB_3253c|M.abscessus_ATCC_199 SSLADETPEGRSIVELA-------AGTIEESVAGEFVAFTASTRMSGLDT
Mflv_2849|M.gilvum_PYR-GCK LENASEHPIAQAVATAATE------------ELGTLPTPEDFANVEGKGV
TH_4361|M.thermoresistible__bu VEHASEHPIARAVATHAAQ------------TVPPLPEVSDFENHGGNGV
MLBr_02000|M.leprae_Br4923 VEAASEHSVATAIVAAYAD------------PRP----VADFVAFAGCGV
MUL_0423|M.ulcerans_Agy99 VESGSEHPIGAAIVAGARE------------RELEVPPATEFTNLAGHGV
. ..* . . ::. * .
Mb1059|M.bovis_AF2122/97 AEVSGIRRIRKGAAAAVMKWVRDHGGH-----PTEEVGAIVDGISSGGGT
Rv1030|M.tuberculosis_H37Rv AEVSGIRRIRKGAAAAVMKWVRDHGGH-----PTEEVGAIVDGISSGGGT
MSMEG_5393|M.smegmatis_MC2_155 GEHR----LRKGAAAAVVDWVRGEGGT-----VPVELFDVVDAISHAGGT
Mvan_4760|M.vanbaalenii_PYR-1 DGHR----LRKGATSAVAEWVRSEGGT-----VPTELGDIVDGISAAGGT
MMAR_0632|M.marinum_M DGHL----LRKGAANAVADWVRTQGGE-----VPPELKATVEAISAGGGT
MAV_1172|M.avium_104 DGHS----LRKGAASSVAEWVRSQRGS-----VPHQLGEIVDGISAGGGT
MAB_3253c|M.abscessus_ATCC_199 VEGR---QIRKGASDAVFTWVASLSGVQEDDPVVVAVRKIADQVASEGGT
Mflv_2849|M.gilvum_PYR-GCK QGIVDGHAVVVGRESLLADWSQH---------LSPELARAKASAEAQGKT
TH_4361|M.thermoresistible__bu SGRVDGHDVRVGR---LAWLGLS---------AGDRLRNAAAQAEAAGHT
MLBr_02000|M.leprae_Br4923 SGVVAEHHVKIGKPS-WVTRNAP---------CDVVLESARREGESRGET
MUL_0423|M.ulcerans_Agy99 RGEVGGKSVLVGRRKLVDEQHLR---------LPEQLAAAAADLEERGRT
: * : * *
Mb1059|M.bovis_AF2122/97 PLVVAEWTDNS----SARAIGVVHLKDIVKVGIRERFDEMRRMSIRTVMI
Rv1030|M.tuberculosis_H37Rv PLVVAEWTDNS----SARAIGVVHLKDIVKVGIRERFDEMRRMSIRTVMI
MSMEG_5393|M.smegmatis_MC2_155 PLVVGHVITDDTDPKRAEVLGVIHLKDVVKQGMKQRFDEMRRMGIRTVMI
Mvan_4760|M.vanbaalenii_PYR-1 PLAVGEVREG-----KAAVLGVIHLKDVVKQGMRERFDEMRRMGIRTVMI
MMAR_0632|M.marinum_M PLAVAQVLDG-----TASVLGVIHLKDVVKHGMRERFDEMRRMGIRTVMI
MAV_1172|M.avium_104 PLVVGESVDG-----RARVLGVIHLKDVVKQGMRERFDEMRRMGIRTVMI
MAB_3253c|M.abscessus_ATCC_199 PLVVADHDST-----ETRLLGVIRLSDVVKPGMAERFAQMRAMGIRTVMV
Mflv_2849|M.gilvum_PYR-GCK AVAVGW---------DGEARGVLVVADTVKPTSAQAISQMREIGLTPVLL
TH_4361|M.thermoresistible__bu PVWVAV---------DGRIAGVIVVSDTVKDHAAEAIAELRALGATPVLL
MLBr_02000|M.leprae_Br4923 VVFVSV---------DGVACGAVAIADTVKDSAADAISALCSRGLHTILL
MUL_0423|M.ulcerans_Agy99 AVYVGQ---------DGQVVGVLAVADTVKDDAADVVSQLHAMGLQVALI
: *. *.: : * ** : . : . ::
Mb1059|M.bovis_AF2122/97 TGDNPATAKAIAQEAGVDDFLAEATPEDKLALIKREQQGGRLVAMTGDGT
Rv1030|M.tuberculosis_H37Rv TGDNPATAKAIAQEAGVDDFLAEATPEDKLALIKREQQGGRLVAMTGDGT
MSMEG_5393|M.smegmatis_MC2_155 TGDNPLTAKAIADEAGVDDFLAEATPEDKLALIKQEQAGGRLVAMTGDGT
Mvan_4760|M.vanbaalenii_PYR-1 TGDNPMTAKAIADEAGVDDFLAEATPEDKMALIKKEQAGGRLVAMTGDGT
MMAR_0632|M.marinum_M TGDNPLTAKAIAEEAGVDDFLAEATPEDKLALIKREQGGGRLVAMTGDGT
MAV_1172|M.avium_104 TGDNPLTAKAIADEAGVDDFLAEATPEDKLQLIKREQAGGKLVAMTGDGT
MAB_3253c|M.abscessus_ATCC_199 TGDNPLTAKQIAAEAGVDDFVAEATPEDKLELLRKEQQAGRLVAMTGDGT
Mflv_2849|M.gilvum_PYR-GCK TGDNEAVARQIAAEVGIDTVIAEVMPKDKVDVIVRLQAEGKTVAMVGDGV
TH_4361|M.thermoresistible__bu TGDNDRAARTVAAQVGIDDVIAGVLPAEKVDHIKQLQDRGRVVAMIGDGV
MLBr_02000|M.leprae_Br4923 TGDNQAAARAVAAQVGIDTVIADMLPEAKVDVIQRLRDQGHTVAMVGDGI
MUL_0423|M.ulcerans_Agy99 TGDNARTAAAIAAQVGIDRVLAEVLPADKVNEVRRLQDEGRIVAMVGDGV
**** .* :* :.*:* .:* * *: : : : *: *** ***
Mb1059|M.bovis_AF2122/97 NDAPALAQADVGVAMNTGTQAAREAGNMVDLDSDPTKLIEVVEIGKQLLI
Rv1030|M.tuberculosis_H37Rv NDAPALAQADVGVAMNTGTQAAREAGNMVDLDSDPTKLIEVVEIGKQLLI
MSMEG_5393|M.smegmatis_MC2_155 NDAPALAQADVGVAMNTGTSAAKEAGNMVDLDSDPTKLIEIVEIGKQLLI
Mvan_4760|M.vanbaalenii_PYR-1 NDAPALAQADVGVAMNSGTSAAKEAGNMVDLDSDPTKLIEIVEIGKQLLI
MMAR_0632|M.marinum_M NDAPALAQADVGVAMNTGTSAAKEAGNMVDLDSDPTKLIEIVEIGKQLLI
MAV_1172|M.avium_104 NDAPALAQADVGVAMNTGTSAAKEAGNMVDLDSDPTKLIEIVEIGKQLLI
MAB_3253c|M.abscessus_ATCC_199 NDAPALAQADVGVAMNTGTSAAKEAGNMVDLDSDPTKLIDVVAIGKQLLI
Mflv_2849|M.gilvum_PYR-GCK NDAPALAQADLGLAMGTGTDVAIEASDITLVRGDLRSAVDAIRLSRRTLS
TH_4361|M.thermoresistible__bu NDAAALAQADLGLAIGTGTDVAIEASDLTLVTGDLRAAPDAIRLGRATLR
MLBr_02000|M.leprae_Br4923 NDGPALACADLGLAMGRGTDVAIGAADLILVRDSLGVVPVALDLARATMR
MUL_0423|M.ulcerans_Agy99 NDAPALVQADLGIAIGTGTDVAIEASDITLMSGRLDGVVSAIELSRRTLR
**..**. **:*:*:. **..* *.:: : . : :.: :
Mb1059|M.bovis_AF2122/97 TRGALTTFSIANDVAKYFAIIPAMFVGLYPVLDKLNVMALHSPRSAILSA
Rv1030|M.tuberculosis_H37Rv TRGALTTFSIANDVAKYFAIIPAMFVGLYPVLDKLNVMALHSPRSAILSA
MSMEG_5393|M.smegmatis_MC2_155 TRGALTTFSISNDIAKYFAIIPALFVGLFPGLDLLNIMRLHSPQSAILSA
Mvan_4760|M.vanbaalenii_PYR-1 TRGALTTFSIANDIAKYFAIIPALFVGLFPGLDLLNVMRLHSPQSAILSA
MMAR_0632|M.marinum_M TRGALTTFSIANDIAKYFAIIPALFVGLFPGLDLLNIMRLHSPQSAILSA
MAV_1172|M.avium_104 TRGALTTFSIANDIAKYFAIIPAMFVALFPGLDLINVMRLHSPQSAILSA
MAB_3253c|M.abscessus_ATCC_199 TRGALTTFSLANDLAKYFAILPALFSGIYPQLGALNIMRLATPQSAILSA
Mflv_2849|M.gilvum_PYR-GCK TIKTNLFWAFV-----------------------YNVAAIPVAALGMLNP
TH_4361|M.thermoresistible__bu TIKGNLFWAFA-----------------------YNVAALPLAAAGLLNP
MLBr_02000|M.leprae_Br4923 TIRINMIWAFG-----------------------YNVAAIPIASSGLLNP
MUL_0423|M.ulcerans_Agy99 TIYQNLGWAFG-----------------------YNTAAIPLAALGVLNP
* ::: * : . .:*..
Mb1059|M.bovis_AF2122/97 VIFNALVIVALIPLALRGVRFRAESASAMLRRNLLIYGLGGLVVPFIGIK
Rv1030|M.tuberculosis_H37Rv VIFNALVIVALIPLALRGVRFRAESASAMLRRNLLIYGLGGLVVPFIGIK
MSMEG_5393|M.smegmatis_MC2_155 VIFNAIVIVALIPLALKGVRYTPSSAAKLLSRNLYIYGLGGIIAPFLGIK
Mvan_4760|M.vanbaalenii_PYR-1 VIFNAVVIIALIPLSLRGVRYTPSSASKLLSRNLYIFGLGGIIAPFVGIK
MMAR_0632|M.marinum_M VIFNALVIVALIPLALRGVRYTPSSASKLLSRNLSIYGLGGIVAPFIGIK
MAV_1172|M.avium_104 VIFNAIIIVLLIPLSLRGVRYTPSSASKLLSRNLYIYGLGGIVAPFIGIK
MAB_3253c|M.abscessus_ATCC_199 VVFNALVIIALVPLALKGVRYRPVGAGELLRRNLLIYGLGGVIVPFVGIW
Mflv_2849|M.gilvum_PYR-GCK MLAGAAMAFSSVFVVGNSLRLRGFTSTSTSSAPTH---------------
TH_4361|M.thermoresistible__bu LIAGAAMAFSSVFVVSNSLRLRRFTPSR----------------------
MLBr_02000|M.leprae_Br4923 LIAGAAMAFSSFFVVSNSLRLSNFGLSQTSD-------------------
MUL_0423|M.ulcerans_Agy99 VVAGAAMGLSSVSVVTNSLRLRRFGRDV----------------------
:: .* : . . : ..:*
Mb1059|M.bovis_AF2122/97 LVDLVIVAL-GVS
Rv1030|M.tuberculosis_H37Rv LVDLVIVAL-GVS
MSMEG_5393|M.smegmatis_MC2_155 LIDLVVQLLPGMS
Mvan_4760|M.vanbaalenii_PYR-1 AIDLIIQLFPGM-
MMAR_0632|M.marinum_M LIDLIIQFLPGMS
MAV_1172|M.avium_104 AIDLIVQFVPGMS
MAB_3253c|M.abscessus_ATCC_199 LIDLVVRFIPGIG
Mflv_2849|M.gilvum_PYR-GCK -------------
TH_4361|M.thermoresistible__bu -------------
MLBr_02000|M.leprae_Br4923 -------------
MUL_0423|M.ulcerans_Agy99 -------------