For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SKSKNAVVTTGVFDPAQLIKALPLAVRKLDPRHMARNPVMFVVTVGSVATTVLAVLHPSIFAWAITAWLW FTVVFANLAEAVAEGRGKAQAASLREVKRDTVARRLVGDGNGNENEEEVPGSALRPGDRVVVEAGQVIPG DGDVVEGIATVDESAITGESAPVVRESGGDRCAVTGGTTVLSDRIVVQITAAPGESFVDRMIALVEGASR QKTPNEIALNILLASLTIIFLLAVVALGPMGNYGGEQQDPIKLIALLVCLIPTTIGALMSAIGIAGMDRL VQHNVLAKSGRAVEAAGDIDTLLMDKTGTITFGNRQATEFLVAPGIPHETLAAAARASSLADETPEGRSI VELAAGTIEESVAGEFVAFTASTRMSGLDTVEGRQIRKGASDAVFTWVASLSGVQEDDPVVVAVRKIADQ VASEGGTPLVVADHDSTETRLLGVIRLSDVVKPGMAERFAQMRAMGIRTVMVTGDNPLTAKQIAAEAGVD DFVAEATPEDKLELLRKEQQAGRLVAMTGDGTNDAPALAQADVGVAMNTGTSAAKEAGNMVDLDSDPTKL IDVVAIGKQLLITRGALTTFSLANDLAKYFAILPALFSGIYPQLGALNIMRLATPQSAILSAVVFNALVI IALVPLALKGVRYRPVGAGELLRRNLLIYGLGGVIVPFVGIWLIDLVVRFIPGIG*
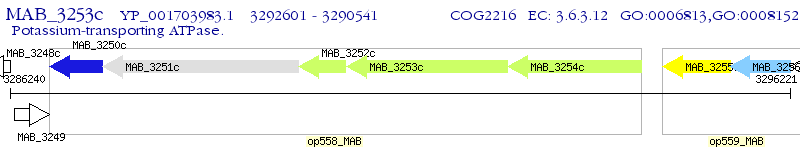
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3253c | - | - | 100% (686) | potassium-transporting ATPase B chain |
| M. abscessus ATCC 19977 | MAB_0449 | - | 3e-51 | 30.02% (523) | copper-transporting ATPase |
| M. abscessus ATCC 19977 | MAB_3984c | - | 4e-44 | 29.65% (516) | putative metal transporter ATPase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1059 | kdpB | 0.0 | 65.99% (685) | potassium-transporting ATPase subunit B |
| M. gilvum PYR-GCK | Mflv_0912 | - | 3e-56 | 31.73% (498) | heavy metal translocating P-type ATPase |
| M. tuberculosis H37Rv | Rv1030 | kdpB | 0.0 | 65.99% (685) | potassium-transporting ATPase subunit B |
| M. leprae Br4923 | MLBr_01987 | ctpA | 2e-52 | 31.85% (496) | putative cation-transporting ATPase |
| M. marinum M | MMAR_0632 | kdpB | 0.0 | 66.33% (701) | high-affinity K+ transport system, ATPase chain B, KdpB |
| M. avium 104 | MAV_1172 | kdpB | 0.0 | 65.71% (700) | potassium-transporting ATPase subunit B |
| M. smegmatis MC2 155 | MSMEG_5393 | kdpB | 0.0 | 67.28% (703) | potassium-transporting ATPase subunit B |
| M. thermoresistible (build 8) | TH_4361 | - | 1e-54 | 32.02% (509) | PUTATIVE heavy metal translocating P-type ATPase |
| M. ulcerans Agy99 | MUL_4850 | ctpA | 8e-55 | 31.56% (526) | cation transporter p-type ATPase a CtpA |
| M. vanbaalenii PYR-1 | Mvan_4760 | - | 0.0 | 67.09% (705) | potassium-transporting ATPase subunit B |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5393|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_4760|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0632|M.marinum_M --------------------------------------------------
MAV_1172|M.avium_104 --------------------------------------------------
Mb1059|M.bovis_AF2122/97 --------------------------------------------------
Rv1030|M.tuberculosis_H37Rv --------------------------------------------------
MAB_3253c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_0912|M.gilvum_PYR-GCK ----MALMTTSTSTPVSGANVELRIGGMTCASCANRIERKLNKLDGVAAT
TH_4361|M.thermoresistible__bu -------MNTGGMNTVT-----LTVGGMTCASCAARVEKRLNRIDGVVAT
MLBr_01987|M.leprae_Br4923 ------------------------------MRAPNGWNNLPNKLSDFSTL
MUL_4850|M.ulcerans_Agy99 MSTTIVTTDKHLERDQQVQRFQLGITGMSCFACARKVQSTLNKVPGVRAS
MSMEG_5393|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_4760|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0632|M.marinum_M --------------------------------------------------
MAV_1172|M.avium_104 --------------------------------------------------
Mb1059|M.bovis_AF2122/97 --------------------------------------------------
Rv1030|M.tuberculosis_H37Rv --------------------------------------------------
MAB_3253c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_0912|M.gilvum_PYR-GCK VNYATEKATVTVPDGYDPALLIAEVEKTGYTATLPTPRAPAPSDASGDTP
TH_4361|M.thermoresistible__bu VNFATEQATVRYPDHVDPADLIAAIEATGYTASLPAPQRDTGGAPAGAAT
MLBr_01987|M.leprae_Br4923 VNSATRVAAINLSEVTQTAEVCEAVRRAALCTDG------------GEAL
MUL_4850|M.ulcerans_Agy99 VNFGTRTAIVEAGEDVDAAALCEVIQQAGYQAEVQP----------DDGQ
MSMEG_5393|M.smegmatis_MC2_155 --------------------------MPVHTAQQAVSNHANPKP-TSQVQ
Mvan_4760|M.vanbaalenii_PYR-1 --------------------------MSVTTAGAAARKHAEPKS-SNKIQ
MMAR_0632|M.marinum_M -------------------------------------------M-STTVG
MAV_1172|M.avium_104 --------------------------MTVTAIDPAEQAHPAPSAGTKRVQ
Mb1059|M.bovis_AF2122/97 -------------------------MMIARMETSATAAAATSAPRLRLAK
Rv1030|M.tuberculosis_H37Rv -------------------------MMIARMETSATAAAATSAPRLRLAK
MAB_3253c|M.abscessus_ATCC_199 ----------------------------------------MSKSKNAVVT
Mflv_0912|M.gilvum_PYR-GCK NADGID-DPELASLRHRLRASTVLTVP----VIAMAMIPALQFTYWQWAS
TH_4361|M.thermoresistible__bu GAAGDDHDAEAERWRRRLTISAALSIP----VVVLSMIPALQFTHWQWLA
MLBr_01987|M.leprae_Br4923 QRRQAD-ADNARYLLIRLAVAAALFVPLAHLSVMFAVLPSTHFPGWEWML
MUL_4850|M.ulcerans_Agy99 GAGDSD-AEHARHLLIRLAVAAVLFVPLADLSVMFAVVPSTRFTGWEWVL
MSMEG_5393|M.smegmatis_MC2_155 GGLLDPKMLWRSLPDALRKLNPRTLWRNPVMFIVEVGAVWSTILAGVNPT
Mvan_4760|M.vanbaalenii_PYR-1 GGLLDPNMLWKSLPSALRKLDPRTMWRNPVMFIVEIGAVWSTILAFLEPS
MMAR_0632|M.marinum_M SGLLDPTMLWKSLPAAVRKLDPRTLWRNPVMFIVELGAVWSTLLAVVDPS
MAV_1172|M.avium_104 GGLLDPKMLWRSTPDALRKLDPRTLWRNPVMFIVEIGAAWSTVLAIVGPT
Mb1059|M.bovis_AF2122/97 RSLFDPMIVRSALPQSLRKLAPRVQARNPVMLVVLVGAVITTLAFLRDLA
Rv1030|M.tuberculosis_H37Rv RSLFDPMIVRSALPQSLRKLAPRVQARNPVMLVVLVGAVITTLAFLRDLA
MAB_3253c|M.abscessus_ATCC_199 TGVFDPAQLIKALPLAVRKLDPRHMARNPVMFVVTVGSVATTVLAVLHPS
Mflv_0912|M.gilvum_PYR-GCK LTLAAPVIVWAAWPFHRAAWANLRHGTATMDTLISLGTVSAFLWSLYALF
TH_4361|M.thermoresistible__bu LTLTSPVVVWGAWPFHRAAVANLRHGTATMDTLISLGVTAAYLWSLWALF
MLBr_01987|M.leprae_Br4923 TALAIPVVTWAAWPFHRVAIHNARYHGASMETLISTGITAATIWSLYTVF
MUL_4850|M.ulcerans_Agy99 TALALPVVTWAAWPFHRIALRNARHHGASMETLISVGITAATIWSLYTVF
: * : * .: :: * . : :
MSMEG_5393|M.smegmatis_MC2_155 ------------------------WFGWLIVAWLWLTVVFANLAEAVAEG
Mvan_4760|M.vanbaalenii_PYR-1 ------------------------WFAWLIVVWLWLTVLFANLAEAVAEG
MMAR_0632|M.marinum_M ------------------------WFAGLIVAWLWLTVIFANLAEAVAEG
MAV_1172|M.avium_104 ------------------------WFAWLTVIWLWLTVLFANLAEAVAEG
Mb1059|M.bovis_AF2122/97 SSTAQEN-----------------VFNGLVAAFLWFTVLFANFAEAMAEG
Rv1030|M.tuberculosis_H37Rv SSTAQEN-----------------VFNGLVAAFLWFTVLFANFAEAMAEG
MAB_3253c|M.abscessus_ATCC_199 ------------------------IFAWAITAWLWFTVVFANLAEAVAEG
Mflv_0912|M.gilvum_PYR-GCK FGTAGTPGMKHPFELTLAPSHGAANIYLEVAAGVTTFILAGRYFEKRSKR
TH_4361|M.thermoresistible__bu FTAAGMPGMTMTFTL-LPADTGEPHIYLEVAAAVTTFIIAGRYFEARAKR
MLBr_01987|M.leprae_Br4923 GHHQSTEHRGVWRAL-----LGSDAIYFEVAAGITVFVLAGKYYTARAKS
MUL_4850|M.ulcerans_Agy99 GSHQLADRKGIWQAL-----LGSDAIYFEVAAGVTVFVLAGRYFEARAKS
: . : :: .. ::
MSMEG_5393|M.smegmatis_MC2_155 RGKAQAESLRKTKADTMARRLT-----ESGAEEAVPAPQLRQGDIVVVEA
Mvan_4760|M.vanbaalenii_PYR-1 RGKAQAESLRKTKTATPARRLTGWAPGTPGREEAVAAALLQPGDIVVVEA
MMAR_0632|M.marinum_M RGKAQADTLRKTKADTMARLLT-----ATGAEEQVTAALLQPGDIVVVDA
MAV_1172|M.avium_104 RGKAQAETLRRAKTQTMARRLRDWAPGSTGIEEAVAATALQQGDIVVVEA
Mb1059|M.bovis_AF2122/97 RGKAQAAALRKVRSETMANRRT-----AAGNIESVPSSRLDLDDVVEVSA
Rv1030|M.tuberculosis_H37Rv RGKAQAAALRKVRSETMANRRT-----AAGNIESVPSSRLDLDDVVEVSA
MAB_3253c|M.abscessus_ATCC_199 RGKAQAASLREVKRDTVARRLVG-DGNGNENEEEVPGSALRPGDRVVVEA
Mflv_0912|M.gilvum_PYR-GCK QAGAALRALLNLGAKDVSVLR-------DGVETKIAIEELMVGDEFVVRP
TH_4361|M.thermoresistible__bu QSGAALRALLDMGANDVAVLR-------DGAEKRVPIGELHVGDVFVVRP
MLBr_01987|M.leprae_Br4923 HASIALLALAALSAKDAAVLQP------DGSEMVIPANELNEQQRFVVRP
MUL_4850|M.ulcerans_Agy99 KAGAALRALAALSAKEVAVLQP------DGSELVVPADELQEQDRFVVRP
:. :* : :. * : . * .
MSMEG_5393|M.smegmatis_MC2_155 GQTIPGDGDVVEGIASVDESAITGESAPVIRESGGDRSAVTGGTTVLSDR
Mvan_4760|M.vanbaalenii_PYR-1 GQVIPGDGDVVEGIASVDESAITGESAPVIRESGGDRSAVTGGTTVLSDR
MMAR_0632|M.marinum_M GQVIPGDGDVVEGIASVDESAITGESAPVIRESGGDRSAVTGGTTVLSDR
MAV_1172|M.avium_104 GQVIPGDGDVVEGIASVDESAITGESAPVIRESGGDRSAVTGGTTVLSDR
Mb1059|M.bovis_AF2122/97 GETIPSDGEIIEGIASVDESAITGESAPVIRESGGDRSAVTGGTVVLSDR
Rv1030|M.tuberculosis_H37Rv GETIPSDGEIIEGIASVDESAITGESAPVIRESGGDRSAVTGGTVVLSDR
MAB_3253c|M.abscessus_ATCC_199 GQVIPGDGDVVEGIATVDESAITGESAPVVRESGGDRCAVTGGTTVLSDR
Mflv_0912|M.gilvum_PYR-GCK GEKIATDGVVTSGSSAVDASMLTGESVPVEVAAGDT---VVGATVNAGGR
TH_4361|M.thermoresistible__bu GEKIATDGEVVAGNSAVDASMLTGESVPVEVGPGDQ---VVGATVNVGGH
MLBr_01987|M.leprae_Br4923 GQTIAADGLVIDGSATVSMSPITGEAKPVRVNPGAQ---VIGGTVVLNGR
MUL_4850|M.ulcerans_Agy99 GQTIAADGLVLDGSAAVDMSTMTGEAKPTRVRPGGQ---VIGGTVVLDGR
*: *. ** : * ::*. * :***: *. .* * *.*. ..:
MSMEG_5393|M.smegmatis_MC2_155 IVVRITQKPGESFIDRMIALVEGANRQKTPNEIALNILLAALTIIFVFAV
Mvan_4760|M.vanbaalenii_PYR-1 IVVKITQKPGESFIDRMIALVEGANRQKTPNEIALNILLASLTIIFVFAV
MMAR_0632|M.marinum_M ILVKITQKPGESFIDKMIALVEGADRQKTPNEIALNILLASLTIIFVFTV
MAV_1172|M.avium_104 IVVQITQKPGESFIDRMIALVEGANRQKTPNEIALNILLAALTIIFVFAV
Mb1059|M.bovis_AF2122/97 IVVRITAKQGQTFIDRMIALVEGAARQQTPNEIALNILLAGLTIIFLLAV
Rv1030|M.tuberculosis_H37Rv IVVRITAKQGQTFIDRMIALVEGAARQQTPNEIALNILLAGLTIIFLLAV
MAB_3253c|M.abscessus_ATCC_199 IVVQITAAPGESFVDRMIALVEGASRQKTPNEIALNILLASLTIIFLLAV
Mflv_0912|M.gilvum_PYR-GCK LVVRASRVGSDTQLAQMAELVERAQTGK-AEVQRLADRISGVFVPIVIAI
TH_4361|M.thermoresistible__bu LQVRATRVGADTQLAQMARLVTEAQSGK-AEVQRLADRVSAVFVPIVIAL
MLBr_01987|M.leprae_Br4923 LIVEAAAVGDETQLAGMVRLVEQAQQQN-ANAQRLADRIASVFVPCVFAV
MUL_4850|M.ulcerans_Agy99 LIVEAAAVGADTRFAGMVRLVEQAQAQK-ADAQRLADRIASVFVPCVFVI
: *. : :: . * ** * : .: * ::.: : ::.:
MSMEG_5393|M.smegmatis_MC2_155 ATLQPLAIYAKANNPGVPDSLALTGNGISGIVMVALLVCLIPTTIGALLS
Mvan_4760|M.vanbaalenii_PYR-1 TTLQPLAIYSKMNNPGVPDSLALNGNGITGIVMVSLLVCLIPTTIGALLS
MMAR_0632|M.marinum_M VTLQPLAIYSKANNPGVADTVALNANGISGIVMVSLLVCLIPTTIGALLS
MAV_1172|M.avium_104 ATLQPLAIYSKVNNPGVPDTQALNTSGVTGIVMVSLLVCLIPTTIGALLS
Mb1059|M.bovis_AF2122/97 VTLQPFAIYS--------------GGGQRVVVLVALLVCLIPTTIGALLS
Rv1030|M.tuberculosis_H37Rv VTLQPFAIYS--------------GGGQRVVVLVALLVCLIPTTIGALLS
MAB_3253c|M.abscessus_ATCC_199 VALGPMGNYG--------------GEQQDPIKLIALLVCLIPTTIGALMS
Mflv_0912|M.gilvum_PYR-GCK AVITLGAWLG-----------AGFSVTAAFTAAVAVLVIACPCALGLATP
TH_4361|M.thermoresistible__bu SALTLAGWLLL----------TDAPVSAAFTAAVAVLIIACPCALGLATP
MLBr_01987|M.leprae_Br4923 AALTAVGWLI-----------AGSGPDRVFSAAIAVLVIACPCALGLATP
MUL_4850|M.ulcerans_Agy99 AALTAAGWLL-----------AGGSPDRAFSAAIAVLVIACPCALGLATP
.: . :::*: * ::* .
MSMEG_5393|M.smegmatis_MC2_155 AIGIAGMDRLVQRNVLAMSGRAVEAAGDVNTLLLDKTGTITLGNRQAAAF
Mvan_4760|M.vanbaalenii_PYR-1 AIGIAGMDRLVQRNVLAMSGRAVEAAGDVNTLLLDKTGTITLGNRQASEF
MMAR_0632|M.marinum_M AIGIAGMDRLVQRNILAMSGRAVEAAGDVNTLLLDKTGTITLGNRQAGAF
MAV_1172|M.avium_104 AIGIAGMDRLVQRNVLAMSGRAVEAAGDVNTLLLDKTGTITLGNRQAAAF
Mb1059|M.bovis_AF2122/97 AIGIAGMDRLVQHNVLATSGRAVEAAGDVNTLLLDKTGTITLGNRQATEF
Rv1030|M.tuberculosis_H37Rv AIGIAGMDRLVQHNVLATSGRAVEAAGDVNTLLLDKTGTITLGNRQATEF
MAB_3253c|M.abscessus_ATCC_199 AIGIAGMDRLVQHNVLAKSGRAVEAAGDIDTLLMDKTGTITFGNRQATEF
Mflv_0912|M.gilvum_PYR-GCK TALLVGTGRGAQMGVLIKGPEVLESTRKVDTVVLDKTGTVTTGKMALVDV
TH_4361|M.thermoresistible__bu TALLVGTGRGAQLGILIKGPQVLESTRRIDTVVLDKTGTVTTGQMSVTSV
MLBr_01987|M.leprae_Br4923 TAMMVASGRGAQLGILLKGHESFEATRAVDTVVFDKTGTLTTGQLKVSAV
MUL_4850|M.ulcerans_Agy99 TAMMVASGRGAQLGIFLKGYQSLEATRSVDTVVFDKTGTLTSGQLKVSAV
: :.. .* .* .:: . . .*:: ::*:::*****:* *: .
MSMEG_5393|M.smegmatis_MC2_155 VPLTGVTPESLADAAQLSSLADETPEGRSIVVFAKREFGLRARTPGELAE
Mvan_4760|M.vanbaalenii_PYR-1 VPLTGVTPEALADAAQLSSLADETPEGRSIVVFAKQEYGLRARTPGELAH
MMAR_0632|M.marinum_M VHLPGVSEAELADAAQLSSLADETPEGRSIVVYAKQQYGLRARTPGELAH
MAV_1172|M.avium_104 IPLAGVPPEELADAAQLSSLADETPEGRSVVVFAKQHFGLRARTPGELSQ
Mb1059|M.bovis_AF2122/97 VPINGVSAEAVADAAQLSSLADETPEGRSIVVLAKDEFGLRARDEGVMSH
Rv1030|M.tuberculosis_H37Rv VPINGVSAEAVADAAQLSSLADETPEGRSIVVLAKDEFGLRARDEGVMSH
MAB_3253c|M.abscessus_ATCC_199 LVAPGIPHETLAAAARASSLADETPEGRSIVELA-------AGTIEESVA
Mflv_0912|M.gilvum_PYR-GCK IAAPGTERAELLRLAGALENASEHPIAHAIATAAAEEVGT-LPTPEGFAN
TH_4361|M.thermoresistible__bu HVADGLSETELLRLAGAVEHASEHPIARAVATHAAQTVPP-LPEVSDFEN
MLBr_01987|M.leprae_Br4923 TAAPGWQANEVLQMAATVESASEHAVALAIAASTTHREP-----VANFRA
MUL_4850|M.ulcerans_Agy99 VAAPGWDADEVLAIAATVEAASEHSVALAIAAATTQHDT-----VTEFRA
* : * . *.* . . ::. :
MSMEG_5393|M.smegmatis_MC2_155 AHWIEFSATTRMSGVNLGEHR----LRKGAAAAVVDWVRGEGGT-----V
Mvan_4760|M.vanbaalenii_PYR-1 AAWIEFSATTRMSGVDVDGHR----LRKGATSAVAEWVRSEGGT-----V
MMAR_0632|M.marinum_M AQWIEFSATTRMSGVNVDGHL----LRKGAANAVADWVRTQGGE-----V
MAV_1172|M.avium_104 AQWVAFSATTRMSGVDLDGHS----LRKGAASSVAEWVRSQRGS-----V
Mb1059|M.bovis_AF2122/97 ARFVPFTAETRMSGVDLAEVSGIRRIRKGAAAAVMKWVRDHGGH-----P
Rv1030|M.tuberculosis_H37Rv ARFVPFTAETRMSGVDLAEVSGIRRIRKGAAAAVMKWVRDHGGH-----P
MAB_3253c|M.abscessus_ATCC_199 GEFVAFTASTRMSGLDTVEGR---QIRKGASDAVFTWVASLSGVQEDDPV
Mflv_0912|M.gilvum_PYR-GCK VEGKGVQGVVDGHAVVVGRES------------LLADWAQHLS-------
TH_4361|M.thermoresistible__bu HGGNGVSGRVDGHDVRVGR---------------LAWLGLSAG-------
MLBr_01987|M.leprae_Br4923 VPGHGVSGTVAERAVRVGK---------------PSWIASRCNS------
MUL_4850|M.ulcerans_Agy99 IPGRGVSGTVSGRSVRVGK---------------PSWIGSSSSD------
. . . : .
MSMEG_5393|M.smegmatis_MC2_155 PVELFDVVDAISHAGGTPLVVGHVITDDTDPKRAEVLGVIHLKDVVKQGM
Mvan_4760|M.vanbaalenii_PYR-1 PTELGDIVDGISAAGGTPLAVGEVREG-----KAAVLGVIHLKDVVKQGM
MMAR_0632|M.marinum_M PPELKATVEAISAGGGTPLAVAQVLDG-----TASVLGVIHLKDVVKHGM
MAV_1172|M.avium_104 PHQLGEIVDGISAGGGTPLVVGESVDG-----RARVLGVIHLKDVVKQGM
Mb1059|M.bovis_AF2122/97 TEEVGAIVDGISSGGGTPLVVAEWTDNS----SARAIGVVHLKDIVKVGI
Rv1030|M.tuberculosis_H37Rv TEEVGAIVDGISSGGGTPLVVAEWTDNS----SARAIGVVHLKDIVKVGI
MAB_3253c|M.abscessus_ATCC_199 VVAVRKIADQVASEGGTPLVVADHDST-----ETRLLGVIRLSDVVKPGM
Mflv_0912|M.gilvum_PYR-GCK -PDLSRAKAGAEAQGKTVVAVGWDGQA---------RGVLVVADTVKPTS
TH_4361|M.thermoresistible__bu -DRLRNAAAQAEAAGHTPVWVAVDGRI---------AGVIVVSDTVKDHA
MLBr_01987|M.leprae_Br4923 -TTLVTARRNAELRGETAVFVEIDGEQ---------CGVIAVADAVKASA
MUL_4850|M.ulcerans_Agy99 -TALRAALRNAESLGETAVFVEIDGGP---------CGVIAVADAVKDSA
: * * : * **: : * **
MSMEG_5393|M.smegmatis_MC2_155 KQRFDEMRRMGIRTVMITGDNPLTAKAIADEAGVDDFLAEATPEDKLALI
Mvan_4760|M.vanbaalenii_PYR-1 RERFDEMRRMGIRTVMITGDNPMTAKAIADEAGVDDFLAEATPEDKMALI
MMAR_0632|M.marinum_M RERFDEMRRMGIRTVMITGDNPLTAKAIAEEAGVDDFLAEATPEDKLALI
MAV_1172|M.avium_104 RERFDEMRRMGIRTVMITGDNPLTAKAIADEAGVDDFLAEATPEDKLQLI
Mb1059|M.bovis_AF2122/97 RERFDEMRRMSIRTVMITGDNPATAKAIAQEAGVDDFLAEATPEDKLALI
Rv1030|M.tuberculosis_H37Rv RERFDEMRRMSIRTVMITGDNPATAKAIAQEAGVDDFLAEATPEDKLALI
MAB_3253c|M.abscessus_ATCC_199 AERFAQMRAMGIRTVMVTGDNPLTAKQIAAEAGVDDFVAEATPEDKLELL
Mflv_0912|M.gilvum_PYR-GCK AQAISQMRDLGLTPVLLTGDNEAVARQIAAEVGIDTVIAEVMPKDKVDVI
TH_4361|M.thermoresistible__bu AEAIAELRALGATPVLLTGDNDRAARTVAAQVGIDDVIAGVLPAEKVDHI
MLBr_01987|M.leprae_Br4923 ADAVAALHDRGFRTALLTGDNPASAAAVASRIGIDEVIADILPEDKVDVI
MUL_4850|M.ulcerans_Agy99 ADAVAALRERGLRTMLLTGDNPASAAAVADRVGIDEVIANILPEGKVDVI
: . :: . . ::**** * :* . *:* .:* * *: :
MSMEG_5393|M.smegmatis_MC2_155 KQEQAGGRLVAMTGDGTNDAPALAQADVGVAMNTGTSAAKEAGNMVDLDS
Mvan_4760|M.vanbaalenii_PYR-1 KKEQAGGRLVAMTGDGTNDAPALAQADVGVAMNSGTSAAKEAGNMVDLDS
MMAR_0632|M.marinum_M KREQGGGRLVAMTGDGTNDAPALAQADVGVAMNTGTSAAKEAGNMVDLDS
MAV_1172|M.avium_104 KREQAGGKLVAMTGDGTNDAPALAQADVGVAMNTGTSAAKEAGNMVDLDS
Mb1059|M.bovis_AF2122/97 KREQQGGRLVAMTGDGTNDAPALAQADVGVAMNTGTQAAREAGNMVDLDS
Rv1030|M.tuberculosis_H37Rv KREQQGGRLVAMTGDGTNDAPALAQADVGVAMNTGTQAAREAGNMVDLDS
MAB_3253c|M.abscessus_ATCC_199 RKEQQAGRLVAMTGDGTNDAPALAQADVGVAMNTGTSAAKEAGNMVDLDS
Mflv_0912|M.gilvum_PYR-GCK ARLQSEGKTVAMVGDGVNDAPALAQADLGLAMGTGTDVAIEASDITLVRG
TH_4361|M.thermoresistible__bu KQLQDRGRVVAMIGDGVNDAAALAQADLGLAIGTGTDVAIEASDLTLVTG
MLBr_01987|M.leprae_Br4923 EQLRDRGHVVAMVGDGINDGPALARADLGMAIGRGTDVAIGAADIILVRD
MUL_4850|M.ulcerans_Agy99 EQLRDRGHVVAMVGDGINDGPALASADLGMAMGRGTDVAIGAADIILVRD
: : *: *** *** **..*** **:*:*:. **..* *.:: : .
MSMEG_5393|M.smegmatis_MC2_155 DPTKLIEIVEIGKQLLITRGALTTFSISNDIAKYFAIIPALFVGLFPGLD
Mvan_4760|M.vanbaalenii_PYR-1 DPTKLIEIVEIGKQLLITRGALTTFSIANDIAKYFAIIPALFVGLFPGLD
MMAR_0632|M.marinum_M DPTKLIEIVEIGKQLLITRGALTTFSIANDIAKYFAIIPALFVGLFPGLD
MAV_1172|M.avium_104 DPTKLIEIVEIGKQLLITRGALTTFSIANDIAKYFAIIPAMFVALFPGLD
Mb1059|M.bovis_AF2122/97 DPTKLIEVVEIGKQLLITRGALTTFSIANDVAKYFAIIPAMFVGLYPVLD
Rv1030|M.tuberculosis_H37Rv DPTKLIEVVEIGKQLLITRGALTTFSIANDVAKYFAIIPAMFVGLYPVLD
MAB_3253c|M.abscessus_ATCC_199 DPTKLIDVVAIGKQLLITRGALTTFSLANDLAKYFAILPALFSGIYPQLG
Mflv_0912|M.gilvum_PYR-GCK DLRSAVDAIRLSRRTLST----IKTNLFWAFAYNVAAIPVAALGMLNPML
TH_4361|M.thermoresistible__bu DLRAAPDAIRLGRATLRT----IKGNLFWAFAYNVAALPLAAAGLLNPLI
MLBr_01987|M.leprae_Br4923 NLDVVPITLDLAAATMRT----IKFNMVWAFGYNIAAIPIAAAGLLNPLV
MUL_4850|M.ulcerans_Agy99 NLDVVPLALGLAAATMRT----VKLNLAWAFGYNIAAIPIAAAGVLNPVV
: : :. : * . .: .. .* :* .: :
MSMEG_5393|M.smegmatis_MC2_155 LLNIMRLHSPQSAILSAVIFNAIVIVALIPLALKGVRYTPSSAAKLLSRN
Mvan_4760|M.vanbaalenii_PYR-1 LLNVMRLHSPQSAILSAVIFNAVVIIALIPLSLRGVRYTPSSASKLLSRN
MMAR_0632|M.marinum_M LLNIMRLHSPQSAILSAVIFNALVIVALIPLALRGVRYTPSSASKLLSRN
MAV_1172|M.avium_104 LINVMRLHSPQSAILSAVIFNAIIIVLLIPLSLRGVRYTPSSASKLLSRN
Mb1059|M.bovis_AF2122/97 KLNVMALHSPRSAILSAVIFNALVIVALIPLALRGVRFRAESASAMLRRN
Rv1030|M.tuberculosis_H37Rv KLNVMALHSPRSAILSAVIFNALVIVALIPLALRGVRFRAESASAMLRRN
MAB_3253c|M.abscessus_ATCC_199 ALNIMRLATPQSAILSAVVFNALVIIALVPLALKGVRYRPVGAGELLRRN
Mflv_0912|M.gilvum_PYR-GCK AGAAMAFSS------VFVVGNSLRLRGFTSTSSAPTH-------------
TH_4361|M.thermoresistible__bu AGAAMAFSS------VFVVSNSLRLRRFTPSR------------------
MLBr_01987|M.leprae_Br4923 AGAAMAFSS------FFVVSNSLRLRNFGAILSCGTSRHRTVKRWRCPPP
MUL_4850|M.ulcerans_Agy99 AGAAMAFSS------FFVVSNSLQLRNFGAATSQTTT-------------
* : : *: *:: : : .
MSMEG_5393|M.smegmatis_MC2_155 LYIYGLGGIIAPFLGIKLIDLVVQLLPGMS-
Mvan_4760|M.vanbaalenii_PYR-1 LYIFGLGGIIAPFVGIKAIDLIIQLFPGM--
MMAR_0632|M.marinum_M LSIYGLGGIVAPFIGIKLIDLIIQFLPGMS-
MAV_1172|M.avium_104 LYIYGLGGIVAPFIGIKAIDLIVQFVPGMS-
Mb1059|M.bovis_AF2122/97 LLIYGLGGLVVPFIGIKLVDLVIVAL-GVS-
Rv1030|M.tuberculosis_H37Rv LLIYGLGGLVVPFIGIKLVDLVIVAL-GVS-
MAB_3253c|M.abscessus_ATCC_199 LLIYGLGGVIVPFVGIWLIDLVVRFIPGIG-
Mflv_0912|M.gilvum_PYR-GCK -------------------------------
TH_4361|M.thermoresistible__bu -------------------------------
MLBr_01987|M.leprae_Br4923 TRLRSTACSPVDASPLRPVAHRTGVKPPTHR
MUL_4850|M.ulcerans_Agy99 -------------STTEPGATE---------