For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PLKIIAEHREPQIGPNTITVAAAQLGGPWLQPAARLTLIAGAAHVAADAGASLIAYPETYLSGYPFWPSR TQGALFDHPDQKRCYAYYLDAAIEIGGPEHREMETLAADLGLTMIVGVTERGRGLGRGTVWCTLLTIDPR RGLVGHHRKLVPTYDERLVWGQGDGAGLVTHPVGKAVVGSLNCWENWMPQARTALYAQGETVHVATWPGS AKLTGDITRFVAAEGRMFTVAASGLVTADSIPDDFPLAAELRQASDTVVFDGGSAIAGPDGQWLIPPLAD EEGVIVAELDLDRVYAERLNFDPTGHYTRPDVFRTVVNRARQQVVRFTEDSFADELRAYP*
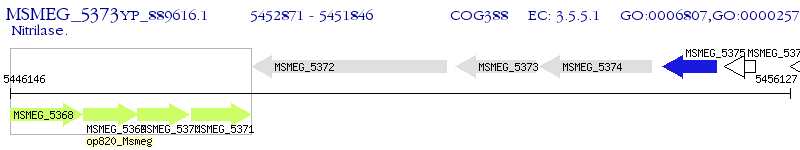
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5373 | - | - | 100% (341) | nitrilase 2 |
| M. smegmatis MC2 155 | MSMEG_1120 | - | 5e-90 | 50.64% (312) | nitrilase/cyanide hydratase and apolipoprotein |
| M. smegmatis MC2 155 | MSMEG_4138 | - | 2e-32 | 30.19% (308) | aliphatic nitrilase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0490c | - | 2e-06 | 24.67% (300) | hypothetical protein Mb0490c |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv0480c | - | 4e-06 | 25.09% (279) | amidohydrolase |
| M. leprae Br4923 | MLBr_02449 | - | 8e-07 | 25.18% (282) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_0819 | - | 9e-10 | 26.67% (285) | carbon-nitrogen hydrolase family protein |
| M. marinum M | MMAR_0805 | - | 2e-06 | 24.82% (282) | amidohydrolase |
| M. avium 104 | MAV_4670 | - | 2e-05 | 24.64% (280) | carbon-nitrogen hydrolase |
| M. thermoresistible (build 8) | TH_0873 | - | 6e-07 | 25.19% (266) | POSSIBLE AMIDOHYDROLASE |
| M. ulcerans Agy99 | MUL_4548 | - | 7e-06 | 24.47% (282) | amidohydrolase |
| M. vanbaalenii PYR-1 | Mvan_1224 | - | 9e-10 | 23.37% (261) | Nitrilase/cyanide hydratase and apolipoprotein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0805|M.marinum_M -------------------------------MRIALAQILS-GTDPTANL
MUL_4548|M.ulcerans_Agy99 -------------------------------MRIALAQILS-GTDPTANL
MLBr_02449|M.leprae_Br4923 -------------------------------MRIALAQILS-GTDPVANL
Mb0490c|M.bovis_AF2122/97 MRRARRRAQAGLPGSCARRCGALVAGPRLARMRIALAQIRS-GTDPAANL
Rv0480c|M.tuberculosis_H37Rv -------------------------------MRIALAQIRS-GTDPAANL
MAV_4670|M.avium_104 -------------------------------MRIALAQILS-GTDPAANL
TH_0873|M.thermoresistible__bu -------------------------------MRIALAQIRS-GTDPSANL
MAB_0819|M.abscessus_ATCC_1997 ---------------------------MTGRLRLAVVQCSSQAADVQANL
Mvan_1224|M.vanbaalenii_PYR-1 -------------------------------MTTLAAVSANFTRDLEQNY
MSMEG_5373|M.smegmatis_MC2_155 -------------MPLKIIAEHREPQIGPNTITVAAAQLGGPWLQPAARL
: . . : .
MMAR_0805|M.marinum_M ELVRDYAERAADAGARLVVFPEATMCRFGV--------------PLGPVA
MUL_4548|M.ulcerans_Agy99 ELVRDYAERAADAGARLVVFPEATMCRFGV--------------PLGPVA
MLBr_02449|M.leprae_Br4923 RLVREYTGQATDAGATLVVFPEATMCRFGV--------------PLGPVA
Mb0490c|M.bovis_AF2122/97 QLVGKYAGEAATAGAQLVVFPEATMCRLGV--------------PLRQVA
Rv0480c|M.tuberculosis_H37Rv QLVGKYAGEAATAGAQLVVFPEATMCRLGV--------------PLRQVA
MAV_4670|M.avium_104 ALVGEYTRRAAGAGARLVVFPEATMCRFGV--------------PLAPIA
TH_0873|M.thermoresistible__bu NLVEEYTRRAAATGAGLVLFPEATMCRFGV--------------SLKPVA
MAB_0819|M.abscessus_ATCC_1997 ARLAETAHAAAAGDAQLVVFPEMYLTGYNIGE-----------WDIRDLA
Mvan_1224|M.vanbaalenii_PYR-1 ALIASLADEARHRGVDFLTLPEAAIGGYLSSLGNHG-------DTVRTTA
MSMEG_5373|M.smegmatis_MC2_155 TLIAGAAHVAADAGASLIAYPETYLSGYPFWPSRTQGALFDHPDQKRCYA
: : * .. :: ** : *
MMAR_0805|M.marinum_M QPL------DGPWANGVRRIAAQTGITVVAGMFTPA---DAGRVSNTLFA
MUL_4548|M.ulcerans_Agy99 QPL------DGPWANGVRRIAAQTGITVVAGMFTPA---DAGRVSNTLFA
MLBr_02449|M.leprae_Br4923 EPI------DGPWANGVRQIATEAGITIIAGIFTPA---DDGRVMNTLIA
Mb0490c|M.bovis_AF2122/97 EPV------DGPWANGVRRIATEAGITVIAGMFTPT---GDGRVTNTLIA
Rv0480c|M.tuberculosis_H37Rv EPV------DGPWANGVRRIATEAGITVIAGMFTPT---GDGRVTNTLIA
MAV_4670|M.avium_104 EPV------DGPWADGVRRIATEANVTVIAGMFTPS---GDGRVKNTLLV
TH_0873|M.thermoresistible__bu EPL------DGPWATAVREIAAGAGVVVVAGMFTPS---GDGRVTNTLLA
MAB_0819|M.abscessus_ATCC_1997 QSP------DGPAMQFIRQTACDADMHICVGYPERA---SDGQIYNSALI
Mvan_1224|M.vanbaalenii_PYR-1 RSLPPAIRLDGPEIARVQQIVG--DLVVAIGFCELA---DDGHTRYNAAA
MSMEG_5373|M.smegmatis_MC2_155 YYLDAAIEIGGPEHREMETLAADLGLTMIVGVTERGRGLGRGTVWCTLLT
.** :. . .: : * . * .
MMAR_0805|M.marinum_M AGPGAPNQPDDHYHKIHLYDAFGFAESRTVAPGHEPVVITVDDVQVGLTV
MUL_4548|M.ulcerans_Agy99 AGPGAPNQPDDHYHKIHLYDAFGFAESRTVAPGHEPVVITVDDFQVGLTV
MLBr_02449|M.leprae_Br4923 AGPGTPNQPDTHYHKIHLYDAFGFTESRTVTAGHEPVVITVDDVQVGLTI
Mb0490c|M.bovis_AF2122/97 AGPGTPNQPDAHYHKIHLYDAFGFTESRTVAPGREPVVVVVDGVRVGLTV
Rv0480c|M.tuberculosis_H37Rv AGPGTPNQPDAHYHKIHLYDAFGFTESRTVAPGREPVVVVVDGVRVGLTV
MAV_4670|M.avium_104 AN--SDDQAVTHYDKIHLYDAFGFTESRTVAPGREPVVVGVDGVRVGLSV
TH_0873|M.thermoresistible__bu TGP----GVAAHYHKIHLYDAYGFTESRTVAPGSEPVTIDVGGVRVGLTT
MAB_0819|M.abscessus_ATCC_1997 CDP--RGRVVLNYRKSHLFGDT--ERNVFTRPGPQLPLAVVHGVSVGLLI
Mvan_1224|M.vanbaalenii_PYR-1 VLDG--STVYGSYRKVHQPLGEGMSYSAGSGYG----VFDTPIGRVGLQI
MSMEG_5373|M.smegmatis_MC2_155 IDP--RRGLVGHHRKLVPTYDERLVWGQGDGAG--LVTHPVGKAVVGSLN
: * . * . **
MMAR_0805|M.marinum_M CYDIRFPALYTELARRGAELIVVSASWGSGPGKLDQ----------WTLL
MUL_4548|M.ulcerans_Agy99 CYDIRFPALYTELARRGAELIVVSASWGSGPGKLDQ----------WTLL
MLBr_02449|M.leprae_Br4923 CYDIRFPALYTELARRGAQLIVVCASWGSGAGKLEQ----------WTLL
Mb0490c|M.bovis_AF2122/97 CYDIRFPALYTELARRGAQLIAVCASWGSGPGKLEQ----------WTLL
Rv0480c|M.tuberculosis_H37Rv CYDIRFPALYTELARRGAQLIAVCASWGSGPGKLEQ----------WTLL
MAV_4670|M.avium_104 CYDIRFPELYTELARRGAQLIAVCASWGAGPGKLDQ----------WTLL
TH_0873|M.thermoresistible__bu CYDIRFPELYVELARRGAELLTVHASWGSGPGKLEQ----------WTLL
MAB_0819|M.abscessus_ATCC_1997 CYDVEFPENVRALAMARADVVVVPTAN--MEPFHAV----------CETV
Mvan_1224|M.vanbaalenii_PYR-1 CYDKAFPEAARIMALRGAQVIASLSAWPAARTATAENLQDDRWTYRFNLF
MSMEG_5373|M.smegmatis_MC2_155 CWENWMPQARTALYAQGETVHVATWPGSAKLTGDITR--------FVAAE
*:: :* : : . .
MMAR_0805|M.marinum_M ARARALDSASYVAAAGQADPGDTASDPGGGSGAPRGVGGSLVASPLGEVV
MUL_4548|M.ulcerans_Agy99 ARARALDSASYVAAAGQADPGDTASDPGGGSGAPRGVGGSLVTSPLGEVV
MLBr_02449|M.leprae_Br4923 ARARALDSMSYVAAVGQADPGGALTDSGPSSAAPTGVGGSLVASPLGEVV
Mb0490c|M.bovis_AF2122/97 ARARALDSMSYVAAAGQADPGDARTGVGASSAAPTGVGGSLVASPLGEVV
Rv0480c|M.tuberculosis_H37Rv ARARALDSMSYVAAAGQADPGDARTGVGASSAAPTGVGGSLVASPLGEVV
MAV_4670|M.avium_104 ARARALDSMSYIAAAGQADPGGSLS----ASGAPTGVGGSLVASPLGEVV
TH_0873|M.thermoresistible__bu ARARALDTTCVVAAVGQAYPGDEIAAQG-----PTGVGGSLVASATGEVL
MAB_0819|M.abscessus_ATCC_1997 VPARAYENQLYVAYANYCGNHDGLAY----------CGGSSIVGPDGTAL
Mvan_1224|M.vanbaalenii_PYR-1 DTARALDNQVFWVASNQSGTFGSLRY----------VGNAKVVDPGGNIL
MSMEG_5373|M.smegmatis_MC2_155 GRMFTVAASGLVTADSIPDDFPLAAELRQASDTVVFDGGSAIAGPDGQWL
: . . *.: :... * :
MMAR_0805|M.marinum_M ASAGSDP-QLVVADIDLAK-VGAAR-----------DNIGVLRNQSRFTQ
MUL_4548|M.ulcerans_Agy99 ASAGSDP-QLVVADIDLAK-VGAAR-----------DNIGMLRNQSRFTQ
MLBr_02449|M.leprae_Br4923 ASAGTES-KLVVADIDVDN-VTTAR-----------ENIAVLRNRSHFA-
Mb0490c|M.bovis_AF2122/97 VSAGTQP-QLLVADIDVDN-VAAAR-----------DRIAVLRNQTDFVQ
Rv0480c|M.tuberculosis_H37Rv VSAGTQP-QLLVADIDVDN-VAAAR-----------DRIAVLRNQTDFVQ
MAV_4670|M.avium_104 ASAGAQP-QLVLADIDVDR-VAQAR-----------KTIAALSNRSDFAQ
TH_0873|M.thermoresistible__bu AAAGPEP-ELLVADLDLDA-ARRAR-----------DTIAVLRNRSDVAQ
MAB_0819|M.abscessus_ATCC_1997 ARAGEGV-GLLVADVDRAV-IDRTR-----------SRHCFLEDRVARAI
Mvan_1224|M.vanbaalenii_PYR-1 ATTLLGS-GMAVAEVDIDETFRTMR-----------AGMFHLRDRRPDVY
MSMEG_5373|M.smegmatis_MC2_155 IPPLADEEGVIVAELDLDRVYAERLNFDPTGHYTRPDVFRTVVNRARQQV
. : :*::* : ::
MMAR_0805|M.marinum_M TDKAQSRR--------
MUL_4548|M.ulcerans_Agy99 TDKAQSRR--------
MLBr_02449|M.leprae_Br4923 ----------------
Mb0490c|M.bovis_AF2122/97 IDKAQSRG--------
Rv0480c|M.tuberculosis_H37Rv IDKAQSRG--------
MAV_4670|M.avium_104 VGRAESVG--------
TH_0873|M.thermoresistible__bu WGKAESVG--------
MAB_0819|M.abscessus_ATCC_1997 SP--------------
Mvan_1224|M.vanbaalenii_PYR-1 GPITEVGAGSDLAHA-
MSMEG_5373|M.smegmatis_MC2_155 VRFTEDSFADELRAYP