For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TVVLVHGNPETDAIWGPLVEALGREGRDDIVRLSPPGFGAPLPDGFSATYLEYRDWLEGELEKFDGPVDL VGHDWGGGHVLNVVMHRPELVRTWVSDVVGIFDPEYVWHDLAQVWQTPGAGEELVDAMMGGTVEDRTAQW AAFLPADIAGALAAAQGPEMGRAILALYRSAADTAAVAEAGGGLEKARAKPGLSVLATEDTYVGSEEIRR RAAERAGARTEVLDGLHHWWMVQDPARAAAMLTRFWATSPR*
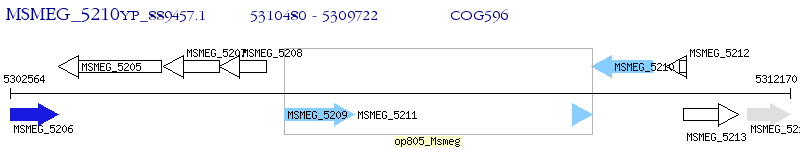
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5210 | - | - | 100% (252) | hydrolase |
| M. smegmatis MC2 155 | MSMEG_6586 | - | 4e-13 | 28.63% (262) | alpha/beta hydrolase, putative |
| M. smegmatis MC2 155 | MSMEG_6719 | - | 2e-05 | 31.91% (188) | epoxide hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0079c | - | 5e-05 | 25.10% (259) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_5155 | - | 1e-102 | 71.37% (248) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | Rv0077c | - | 5e-05 | 25.10% (259) | oxidoreductase |
| M. leprae Br4923 | MLBr_00862 | ephD | 5e-05 | 34.18% (79) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0768 | - | 7e-06 | 25.44% (228) | hypothetical protein MAB_0768 |
| M. marinum M | MMAR_0112 | dhaA_1 | 3e-05 | 28.10% (121) | haloalkane dehalogenase DhaA_1 |
| M. avium 104 | MAV_4567 | - | 1e-07 | 22.35% (264) | dihydrolipoyllysine-residue acetyltransferase component of |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1175 | - | 1e-103 | 71.20% (250) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5155|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1175|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5210|M.smegmatis_MC2_155 --------------------------------------------------
Mb0079c|M.bovis_AF2122/97 ----------------------------------------MSTIDISAGT
Rv0077c|M.tuberculosis_H37Rv ----------------------------------------MSTIDISAGT
MAB_0768|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_4567|M.avium_104 ---------------------------------------MSELKFLELHG
MMAR_0112|M.marinum_M MPAGAPPIDCRYGLPGHYARVQAFSPTHLEAPMTEIDPTPLTKRRLAVDG
MLBr_00862|M.leprae_Br4923 ----------------------------MPATQSIHKVPHQFVESPDGVR
Mflv_5155|M.gilvum_PYR-GCK -----------MTVVFVHGNPETAAIWGPLTAVLGR---DDVVLLSPPGF
Mvan_1175|M.vanbaalenii_PYR-1 -----------MTLVFVHGNPETDAIWGPLTAVLGR---DDVVLLSPPGF
MSMEG_5210|M.smegmatis_MC2_155 -----------MTVVLVHGNPETDAIWGPLVEALGREGRDDIVRLSPPGF
Mb0079c|M.bovis_AF2122/97 IHYEATGPETGRPVVFVHGYMMGGQLWRRVSERLAGRGLRCIAPTWPLGA
Rv0077c|M.tuberculosis_H37Rv IHYEATGPETGRPVVFVHGYMMGGQLWRRVSERLAGRGLRCIAPTWPLGA
MAB_0768|M.abscessus_ATCC_1997 -------MSSKPTIVLVHGFWGGAAHWGNVIVELHGRGFEDLHAVEN---
MAV_4567|M.avium_104 DRVAFRDQGEGEVLLLIHGMAGSSETWRSVIPPLAKK--FRVIAPDLLGH
MMAR_0112|M.marinum_M KQMAYHESGEGRSVVFLHGNPTSSYLWRNIIPHVSDR--ARCIAPDLIGQ
MLBr_00862|M.leprae_Br4923 LAIYQSGQPEGPAIVLVHGFPDSHVLWDGVVPLLAKR--FRIVRYDNRGV
::::** * : :
Mflv_5155|M.gilvum_PYR-GCK G-APLPDGFG---ATFVEYRDWLEAELEQIDG--PIDLVGHDWGGGHVVN
Mvan_1175|M.vanbaalenii_PYR-1 G-TPLPDGFG---ATMLEYRDWLEAELERIDG--PIDLVGHDWGGGHVVN
MSMEG_5210|M.smegmatis_MC2_155 G-APLPDGFS---ATYLEYRDWLEGELEKFDG--PVDLVGHDWGGGHVLN
Mb0079c|M.bovis_AF2122/97 HPKPLRPGAD---QTIGGVAGIVADVLAALELK-DVVLVGNDTGGVVTQL
Rv0077c|M.tuberculosis_H37Rv HPKPLRPGAD---QTIGGVAGIVADVLAALELK-DVVLVGNDTGGVVTQL
MAB_0768|M.abscessus_ATCC_1997 ---PLTS--------LADDAARTRQMVQQIDG--PVLLVGHSYGG----A
MAV_4567|M.avium_104 G-ESAKPRTD---YSLGAFAVWLRDFLDELGVS-RATVVGHSLGGGVAMQ
MMAR_0112|M.marinum_M GDSDKLDDTGPGSYRFVEHRHYLDGLLDQLDLGDDVVLVIHDWGSALGFD
MLBr_00862|M.leprae_Br4923 GQSSVPKPVS--AYTMTRFADDFAAVIDELSPDRPVHVLAHDWGSVGVWE
: : :: :. *.
Mflv_5155|M.gilvum_PYR-GCK AVMHRPELVR----------------------------------------
Mvan_1175|M.vanbaalenii_PYR-1 VVMHRPELVR----------------------------------------
MSMEG_5210|M.smegmatis_MC2_155 VVMHRPELVR----------------------------------------
Mb0079c|M.bovis_AF2122/97 VAVHYPERLG----------------------------------------
Rv0077c|M.tuberculosis_H37Rv VAVHYPERLG----------------------------------------
MAB_0768|M.abscessus_ATCC_1997 VITEAGDLPN----------------------------------------
MAV_4567|M.avium_104 FVYQHPDYAQ----------------------------------------
MMAR_0112|M.marinum_M WANRHRERIGGI--------------------------------------
MLBr_00862|M.leprae_Br4923 YLSRSGASDRVASFTSVSGPSQDQLVDYILSGLRRPWRPRTFARAVSQLF
.
Mflv_5155|M.gilvum_PYR-GCK --------------------------------------------SWASDV
Mvan_1175|M.vanbaalenii_PYR-1 --------------------------------------------SWVSDV
MSMEG_5210|M.smegmatis_MC2_155 --------------------------------------------TWVSDV
Mb0079c|M.bovis_AF2122/97 --------------------------------------------ALVLTS
Rv0077c|M.tuberculosis_H37Rv --------------------------------------------ALVLTS
MAB_0768|M.abscessus_ATCC_1997 --------------------------------------------VAGLVY
MAV_4567|M.avium_104 --------------------------------------------RLILIS
MMAR_0112|M.marinum_M ------------------------------------------AFMEAIVR
MLBr_00862|M.leprae_Br4923 RLTYMVLFSIPVLVPMVLWIALSSATIRRTMVDNISTDQIHHSATLARDA
Mflv_5155|M.gilvum_PYR-GCK VGLFD--PDYQWHDMAQVWQTPGAGEELIANMMGGGLEAR----------
Mvan_1175|M.vanbaalenii_PYR-1 VGLFD--PEYVWHDMAQVWQTPGAGEESLDAMLGGGLTAR----------
MSMEG_5210|M.smegmatis_MC2_155 VGIFD--PEYVWHDLAQVWQTPGAGEELVDAMMGGTVEDR----------
Mb0079c|M.bovis_AF2122/97 CDAFEHFPPPILKPVILAAKSATLFRAAIQVMRAPAARNR----------
Rv0077c|M.tuberculosis_H37Rv CDAFEHFPPPILKPVILAAKSATLFRAAIQVMRAPAARNR----------
MAB_0768|M.abscessus_ATCC_1997 VAAFAP----------DAGESPGQLTEQLPPAAAANLVP-----------
MAV_4567|M.avium_104 SGGLGPDVGWVLRLLSAPGAELVLPVIAPTPVLSVGNKLRSW--------
MMAR_0112|M.marinum_M PVTWAEWPESAAGIFRGFRSDAGEQMVIDKNLFVEAVLPG----------
MLBr_00862|M.leprae_Br4923 AHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQLIVNTMDRYVRPYGYDAT
Mflv_5155|M.gilvum_PYR-GCK ------------TELMSSLGIPADIAAAIAEHQN--DDMGRAILALYRSA
Mvan_1175|M.vanbaalenii_PYR-1 ------------TELMASFGIPADIGAAIAEHQN--DDMRTAILALYRSA
MSMEG_5210|M.smegmatis_MC2_155 ------------TAQWAAF-LPADIAGALAAAQG--PEMGRAILALYRSA
Mb0079c|M.bovis_AF2122/97 ------------AYAGLSHHNIDHLTRAWVRPAL--SNPAIAEDLRQLSL
Rv0077c|M.tuberculosis_H37Rv ------------AYAGLSHHNIDHLTRAWVRPAL--SNPAIAEDLRQLSL
MAB_0768|M.abscessus_ATCC_1997 ------------DSDGYLWIKQDKFRESFAQDLP--EDAALVMAVTQKAP
MAV_4567|M.avium_104 -----------LRGAGIQSPRGAELWNAYSSLSD--GETRQSFLKTLRSV
MMAR_0112|M.marinum_M -----------SILRGLTPAEHDEYRRPFVEPRHRRPTLSWPREIPIEGE
MLBr_00862|M.leprae_Br4923 ARWVPRLWRRDIKAGHFSPMSHPQVMAAAVHDLADLVDGKQPSRALLRAQ
. . .
Mflv_5155|M.gilvum_PYR-GCK TQP------AMAEAGRSLEAAAARPGLSLMATEDPYVGVNDNRRRAADRA
Mvan_1175|M.vanbaalenii_PYR-1 AQP------AMAEAGRALESAAARPGLALLATDDPFIGANDNRRRAAERA
MSMEG_5210|M.smegmatis_MC2_155 ADTA-----AVAEAGGGLEKARAKPGLSVLATEDTYVGSEEIRRRAAERA
Mb0079c|M.bovis_AF2122/97 SLRT-----EVTTAVAARLPEFDKPALIAWSADDVFFALENGQRLAATIP
Rv0077c|M.tuberculosis_H37Rv SLRT-----EVTTAVAARLPEFDKPALIAWSADDVFFALENGQRLAATIP
MAB_0768|M.abscessus_ATCC_1997 LAS-------TFGDAITAPAWRNKPSWYQVSTQDRMINPENERRMAQRIK
MAV_4567|M.avium_104 VDYRG----QAVSALNRLRLREELPVMAIWGERDGIIPVDHAYAAHEART
MMAR_0112|M.marinum_M PADVH----QIASDYAQWLPTADFPKLFINAEPGAILTGP--QREFCRSW
MLBr_00862|M.leprae_Br4923 VGRSRDTFGDTLVSVTGAGSGIGRETALALARRGAEVVVSDIDEAAAKNT
. . .
Mflv_5155|M.gilvum_PYR-GCK GARTTVLDGLGHWWMVED-PQRGAAALTEFWDSL----------------
Mvan_1175|M.vanbaalenii_PYR-1 GARVAVLDGLGHWWMVED-PHRGAEALTEFWATLG---------------
MSMEG_5210|M.smegmatis_MC2_155 GARTEVLDGLHHWWMVQD-PARAAAMLTRFWATSPR--------------
Mb0079c|M.bovis_AF2122/97 RARFEVIEGARTFSMVDS-PDRLADQLSTVAVRT----------------
Rv0077c|M.tuberculosis_H37Rv RARFEVIEGARTFSMVDS-PDRLADQLSTVAVRT----------------
MAB_0768|M.abscessus_ATCC_1997 PRKTIELD-ASHASLASQ-PVAIADLIAEAAGDIA---------------
MAV_4567|M.avium_104 DARLEVLPDVGHFAQVEA-PMRVVELIEDFIATGERRDATSPQPG-----
MMAR_0112|M.marinum_M TNLTEVTVPGSHFIQEDS-PHEIGQAIADWLSSLG---------------
MLBr_00862|M.leprae_Br4923 AAQIVASGGVAYAYALDVSDAAAVEAFAEQVSAKHGVPDIVVNNAGIGQA
. :
Mflv_5155|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1175|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5210|M.smegmatis_MC2_155 --------------------------------------------------
Mb0079c|M.bovis_AF2122/97 --------------------------------------------------
Rv0077c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_0768|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_4567|M.avium_104 --------------------------------------------------
MMAR_0112|M.marinum_M --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GRFLDTPPEEFDRVLAVNLGGVVNCCRAFGQRLVERGTGGHIVNVSSMAA
Mflv_5155|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1175|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5210|M.smegmatis_MC2_155 --------------------------------------------------
Mb0079c|M.bovis_AF2122/97 --------------------------------------------------
Rv0077c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_0768|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_4567|M.avium_104 --------------------------------------------------
MMAR_0112|M.marinum_M --------------------------------------------------
MLBr_00862|M.leprae_Br4923 YAPLQSLSAYCTSKAATYMFSDCLRAELDAVGVGLTTICPGVIDTNIIQS
Mflv_5155|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1175|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5210|M.smegmatis_MC2_155 --------------------------------------------------
Mb0079c|M.bovis_AF2122/97 --------------------------------------------------
Rv0077c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_0768|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_4567|M.avium_104 --------------------------------------------------
MMAR_0112|M.marinum_M --------------------------------------------------
MLBr_00862|M.leprae_Br4923 TRIDTPTAKQERVRDRRGQLDMMFRLRRYGPDKVADTILSAIKKNKPIRP
Mflv_5155|M.gilvum_PYR-GCK ----------------------------
Mvan_1175|M.vanbaalenii_PYR-1 ----------------------------
MSMEG_5210|M.smegmatis_MC2_155 ----------------------------
Mb0079c|M.bovis_AF2122/97 ----------------------------
Rv0077c|M.tuberculosis_H37Rv ----------------------------
MAB_0768|M.abscessus_ATCC_1997 ----------------------------
MAV_4567|M.avium_104 ----------------------------
MMAR_0112|M.marinum_M ----------------------------
MLBr_00862|M.leprae_Br4923 VAPEAYALYGISRVLPQVLRSTARIRVI