For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TVVFVHGNPETAAIWGPLTAVLGRDDVVLLSPPGFGAPLPDGFGATFVEYRDWLEAELEQIDGPIDLVGH DWGGGHVVNAVMHRPELVRSWASDVVGLFDPDYQWHDMAQVWQTPGAGEELIANMMGGGLEARTELMSSL GIPADIAAAIAEHQNDDMGRAILALYRSATQPAMAEAGRSLEAAAARPGLSLMATEDPYVGVNDNRRRAA DRAGARTTVLDGLGHWWMVEDPQRGAAALTEFWDSL*
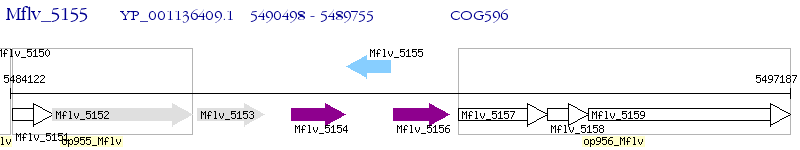
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_5155 | - | - | 100% (247) | alpha/beta hydrolase fold |
| M. gilvum PYR-GCK | Mflv_2400 | - | 4e-05 | 26.49% (151) | alpha/beta hydrolase fold |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2946c | - | 5e-07 | 22.09% (258) | Alpha/beta hydrolase fold |
| M. marinum M | MMAR_0112 | dhaA_1 | 3e-05 | 27.87% (122) | haloalkane dehalogenase DhaA_1 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_5210 | - | 1e-102 | 71.37% (248) | hydrolase |
| M. thermoresistible (build 8) | TH_3479 | - | 9e-05 | 24.02% (254) | PUTATIVE alpha/beta hydrolase fold |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1175 | - | 1e-127 | 85.02% (247) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5155|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1175|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5210|M.smegmatis_MC2_155 --------------------------------------------------
TH_3479|M.thermoresistible__bu ----------------------------------------MSATTITVDD
MMAR_0112|M.marinum_M MPAGAPPIDCRYGLPGHYARVQAFSPTHLEAPMTEIDPTPLTKRRLAVDG
MAB_2946c|M.abscessus_ATCC_199 --------------------------------------MIAESRAEYGGV
Mflv_5155|M.gilvum_PYR-GCK -----------MTVVFVHGNPETAAIWGPLTAVLGR---DDVVLLSPPGF
Mvan_1175|M.vanbaalenii_PYR-1 -----------MTLVFVHGNPETDAIWGPLTAVLGR---DDVVLLSPPGF
MSMEG_5210|M.smegmatis_MC2_155 -----------MTVVLVHGNPETDAIWGPLVEALGREGRDDIVRLSPPGF
TH_3479|M.thermoresistible__bu AAIEYTDVGGGPPIVFVHGAYVTGALWDDVVTRLSRG-HRCIAPTWPFGA
MMAR_0112|M.marinum_M KQMAYHESGEGRSVVFLHGNPTSSYLWRNIIPHVSDR--ARCIAPDLIGQ
MAB_2946c|M.abscessus_ATCC_199 VTRVLSVPGGGPPIVLLHGYSDSALTWRPVLARLASAG-RAAIAVDLPGF
.:*::** : * : :. *
Mflv_5155|M.gilvum_PYR-GCK G-APLPDGFG---ATFVEYRDWLEAELEQIDG--PIDLVGHDWGGGHVVN
Mvan_1175|M.vanbaalenii_PYR-1 G-TPLPDGFG---ATMLEYRDWLEAELERIDG--PIDLVGHDWGGGHVVN
MSMEG_5210|M.smegmatis_MC2_155 G-APLPDGFS---ATYLEYRDWLEGELEKFDG--PVDLVGHDWGGGHVLN
TH_3479|M.thermoresistible__bu QRTPVGDSVD---LDVVACGRRIIGLLEQLDLR-DVTLVANDSGGGITLS
MMAR_0112|M.marinum_M GDSDKLDDTGPGSYRFVEHRHYLDGLLDQLDLGDDVVLVIHDWGSALGFD
MAB_2946c|M.abscessus_ATCC_199 G-HAAPRPPG---PLLAQFDVFADAVLAAAGP---AVLVGNSLGAATAVR
. . * . ** :. *.. .
Mflv_5155|M.gilvum_PYR-GCK AV----MHRPELVRSWAS--DVVGLFDPDYQWHDMAQVWQTPGAGEELIA
Mvan_1175|M.vanbaalenii_PYR-1 VV----MHRPELVRSWVS--DVVGLFDPEYVWHDMAQVWQTPGAGEESLD
MSMEG_5210|M.smegmatis_MC2_155 VV----MHRPELVRTWVS--DVVGIFDPEYVWHDLAQVWQTPGAGEELVD
TH_3479|M.thermoresistible__bu ALGILGLDWSRVSRLVFTNCDTFEHFPPKQFAPLVKLCRVAPPLGGAVLR
MMAR_0112|M.marinum_M WAN---RHRERIGGIAFMEAIVRPVTWAEWPESAAGIFRGFRSDAGEQMV
MAB_2946c|M.abscessus_ATCC_199 AAARRENEVKALIALDDPLNSRHLIARIVRRWSVPEVFWAWVSRMPVPAG
. :
Mflv_5155|M.gilvum_PYR-GCK NMMGG-GLEARTELMSSLGIPADIAAAIAEHQNDDMGRAILALYRSATQP
Mvan_1175|M.vanbaalenii_PYR-1 AMLGG-GLTARTELMASFGIPADIGAAIAEHQNDDMRTAILALYRSAAQP
MSMEG_5210|M.smegmatis_MC2_155 AMMGG-TVEDRTAQWAAF-LPADIAGALAAAQGPEMGRAILALYRSAADT
TH_3479|M.thermoresistible__bu LLATGPGLAFFKRAVTRNGITGDRDDAIFGGFLSSAAVRREAVRFSAGLH
MMAR_0112|M.marinum_M IDKNLFVEAVLPGSILRGLTPAEHDEYRRPFVEPRHRRPTLSWPREIPIE
MAB_2946c|M.abscessus_ATCC_199 TLRRA-ARYFVPKMLYGPGVAADPEVVAYWCELISGPSAVARLGRDACRY
..: .
Mflv_5155|M.gilvum_PYR-GCK -------AMAEAGRSLEAAAARPGLSLMATEDPYVGVNDNRRRAADRAGA
Mvan_1175|M.vanbaalenii_PYR-1 -------AMAEAGRALESAAARPGLALLATDDPFIGANDNRRRAAERAGA
MSMEG_5210|M.smegmatis_MC2_155 A------AVAEAGGGLEKARAKPGLSVLATEDTYVGSEEIRRRAAERAGA
TH_3479|M.thermoresistible__bu P------QHTAAAAQAIGKWTKPVLVLWGTADDLFPVSHAQRLAEAFPNS
MMAR_0112|M.marinum_M GEPADVHQIASDYAQWLPTADFPKLFINAEPGAILTGPQ-REFCRSWTNL
MAB_2946c|M.abscessus_ATCC_199 -------AHEALGSHSGIEVSCPTLVVHGGRDRIIPIHSSGALQQQIPGS
* * : . . . ..
Mflv_5155|M.gilvum_PYR-GCK RTTVLDGLGHWWMVEDPQRGAAALTEFWDSL------------
Mvan_1175|M.vanbaalenii_PYR-1 RVAVLDGLGHWWMVEDPHRGAEALTEFWATLG-----------
MSMEG_5210|M.smegmatis_MC2_155 RTEVLDGLHHWWMVQDPARAAAMLTRFWATSPR----------
TH_3479|M.thermoresistible__bu RLRLIEGSSTYVMLDHPSETADAIGTFIAGRS-----------
MMAR_0112|M.marinum_M TEVTVPG-SHFIQEDSPHEIGQAIADWLSSLG-----------
MAB_2946c|M.abscessus_ATCC_199 ELVILPASGHCPQLDNPDEVTRLIVDFCDRISEPRQPDAAHSG
: . : * . : :