For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTNDVAIIGVGLHPFGRFDKPAVQMGAEAIRLALKDAGVEWKDIQFGFGGSYEVSNPDSVTRLVGLTGIT FTNVFNACATSASAIQQTADTIRLGKYDIGVAVGLDKHPRGAFTDDPAKLALPQWYAQNGQFVTTKFFGM KANKYLHDHNISQDTLARVANKNFRNGALNPNAFRRKEISVEEILASPTLNHPLTQYMFCAPDEGAAAVI MCRADIAHKFTDKPVYVRASEIRTRTFGAYEVHATSAPPESDAAPTVYAARAAYEAAGIGPEDVDIAQLQ DTDAGAEVIHMAETGLCADGDQEKLIAEGATEIDGSIPVNTDGGLIANGEPIGASGLRQMHELVRQLRGE AGDRQVPGNPRVGLAQVYGAPGTASATILSL
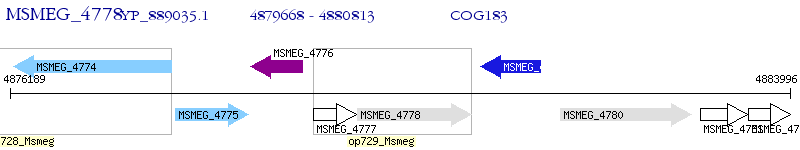
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4778 | - | - | 100% (381) | putative thiolase |
| M. smegmatis MC2 155 | MSMEG_2236 | - | e-171 | 75.07% (381) | putative thiolase |
| M. smegmatis MC2 155 | MSMEG_3844 | - | 3e-50 | 33.42% (395) | lipid-transfer protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1653c | - | 4e-50 | 32.66% (395) | lipid-transfer protein |
| M. gilvum PYR-GCK | Mflv_2519 | - | 0.0 | 90.24% (379) | thiolase |
| M. tuberculosis H37Rv | Rv1627c | - | 4e-50 | 32.66% (395) | lipid-transfer protein |
| M. leprae Br4923 | MLBr_02158 | - | 2e-24 | 54.39% (114) | hypothetical protein MLBr_02158 |
| M. abscessus ATCC 19977 | MAB_2619 | - | 2e-54 | 33.92% (395) | lipid-transfer protein |
| M. marinum M | MMAR_2430 | - | 2e-49 | 33.33% (393) | nonspecific lipid-transfer protein |
| M. avium 104 | MAV_0989 | - | 0.0 | 90.81% (381) | putative thiolase |
| M. thermoresistible (build 8) | TH_0532 | - | 0.0 | 88.36% (378) | putative thiolase |
| M. ulcerans Agy99 | MUL_4955 | ltp1_1 | 4e-46 | 32.17% (401) | lipid-transfer protein Ltp1_1 |
| M. vanbaalenii PYR-1 | Mvan_4139 | - | 0.0 | 90.21% (378) | thiolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1653c|M.bovis_AF2122/97 --MRMS-APEPVYILGAGMHPWGKW-GNDFTEYGVVAARAALRDAGVDWR
Rv1627c|M.tuberculosis_H37Rv --MRMS-APEPVYILGAGMHPWGKW-GNDFTEYGVVAARAALRDAGVDWR
MMAR_2430|M.marinum_M ----MS-APEPLYILGAGMHPWGKW-GNDFTEYGVAAARAALREAGLEWR
MAB_2619|M.abscessus_ATCC_1997 ----MSRTPDPVYILGAGMHPWGKW-GRDFTEYGVAAARTALQDAELDWR
MSMEG_4778|M.smegmatis_MC2_155 -------MTNDVAIIGVGLHPFGRF-DKPAVQMGAEAIRLALKDAGVEWK
MAV_0989|M.avium_104 -------MTNDVAIIGVGLHPFGRF-DKTAMQMGAEAVQLALADAGVAWK
Mflv_2519|M.gilvum_PYR-GCK MTSS----ANDVAIIGVGLHPFGRF-DKTAMEMGAEAIQSALTDAGVDWK
Mvan_4139|M.vanbaalenii_PYR-1 MTNSSPNAAGEVAIIGVGLHPFGRF-DKTAMQMGAEAIQSALTDAGVEWK
TH_0532|M.thermoresistible__bu -----MSNENSVAIIGVGLHPFGRF-DKTAMEMGAEAIAAALADAGLEWK
MLBr_02158|M.leprae_Br4923 --------------------------------------------------
MUL_4955|M.ulcerans_Agy99 -----MSASDEIWILGIRMTKFGKHPDLDTVDLAAQAAMSALDDAGVSMA
Mb1653c|M.bovis_AF2122/97 HVQLVAGADTIRNGYPGFVAGATFAQKLGWTGVPVSSSYAACASGSQALQ
Rv1627c|M.tuberculosis_H37Rv HVQLVAGADTIRNGYPGFVAGATFAQKLGWTGVPVSSSYAACASGSQALQ
MMAR_2430|M.marinum_M QIQLVAGADTIRNGYPGFVAGATFAQKLGWNGVPVSSSYAACASGSQALQ
MAB_2619|M.abscessus_ATCC_1997 QIQFVAGADTIRNGYPGFIAGSTFAQKLGWNGVPISSTYAACASGSQALQ
MSMEG_4778|M.smegmatis_MC2_155 DIQFGFGGSYEVS------NPDSVTRLVGLTGITFTNVFNACATSASAIQ
MAV_0989|M.avium_104 DIQFGFGGSYEVS------NPDAVTRLVGLTGITFTNVFNACATAASAIQ
Mflv_2519|M.gilvum_PYR-GCK DIQFGFGGSYEVS------NPDAVTRLVGLTGITFTNVFNACATAASAIQ
Mvan_4139|M.vanbaalenii_PYR-1 DIQFGFGGSYEVS------NPDAVTRLVGLTGITFTNVFNACATAASAIQ
TH_0532|M.thermoresistible__bu DIQFGYGGSYEIS------NPDAVTRLVGLTGITFTNVFNACATAASAIQ
MLBr_02158|M.leprae_Br4923 --------------------------------------------------
MUL_4955|M.ulcerans_Agy99 DIGVLAAGNLMAANAG---IGQQLQKQIGQTGIPVYNVANACATGATALR
Mb1653c|M.bovis_AF2122/97 SARAQILAGFCDVALVIGADTTPKGFFAPVGGERKGDPDWQRFH------
Rv1627c|M.tuberculosis_H37Rv SARAQILAGFCDVALVIGADTTPKGFFAPVGGERKGDPDWQRFH------
MMAR_2430|M.marinum_M SARAQILAGFCDVALVVGADTTPKGFFAPVGGERKNDPDWQRFH------
MAB_2619|M.abscessus_ATCC_1997 SARAQILAGFCDVALVIGADTTPKGFFAPVGGERKADPDWQRFH------
MSMEG_4778|M.smegmatis_MC2_155 QTADTIRLGKYDIGVAVGLDKHPRGAFTDDPAKLALPQWYAQNG------
MAV_0989|M.avium_104 QTADTIRLGKYDLGIAIGLDKHPRGAFTDDPAKLALPQWYANNG------
Mflv_2519|M.gilvum_PYR-GCK QTADTIRLGKYDIGIAIGLDKHPRGAFTDDPAKLALPQWYANNG------
Mvan_4139|M.vanbaalenii_PYR-1 QTADTIRLGKYDIGIAIGLDKHPRGAFTDDPAKLALPQWYAENG------
TH_0532|M.thermoresistible__bu ETADTIRLGKYDIGIAIGMDKHPRGAFTDDPAKLALPQWYAQNG------
MLBr_02158|M.leprae_Br4923 --------------------------------------------------
MUL_4955|M.ulcerans_Agy99 TAIMAVKAGEVDYGMAVGVEKLSGAGLLGGGGKKKSDADVWTPAGRYGAV
Mb1653c|M.bovis_AF2122/97 ------LIGATNTVYFALLARRRMDLYG-ATVEDFAQVKVKNSRHGLDNP
Rv1627c|M.tuberculosis_H37Rv ------LIGATNTVYFALLARRRMDLYG-ATVEDFAQVKVKNSRHGLDNP
MMAR_2430|M.marinum_M ------LIGATNPVYFALLARRRMDLYG-ATLEDFAQVKVKNSRHGLQNP
MAB_2619|M.abscessus_ATCC_1997 ------LIGATNPVYFALLARRRMDLFG-ATVEDFAQVKVKNSRHGLNNP
MSMEG_4778|M.smegmatis_MC2_155 ------QFVTT--KFFGMKANKYLHDHN-ISQDTLARVANKNFRNGALNP
MAV_0989|M.avium_104 ------QFVTT--KFFGMKANKYIHDHN-ISQETLARVANKNFRNGALNP
Mflv_2519|M.gilvum_PYR-GCK ------QFVTT--KFFGMKANHYLHKHN-ISEETLARVANKNFRNGVINP
Mvan_4139|M.vanbaalenii_PYR-1 ------QFVTT--KFFGMKANHYIHKHN-ISEETLARVANKNFRNGVLNP
TH_0532|M.thermoresistible__bu ------QFVTT--KFFGMKANRYLHDHG-ISQETLARVANKNFRNGSLNP
MLBr_02158|M.leprae_Br4923 ----------------------------------MCSVASRFLR------
MUL_4955|M.ulcerans_Agy99 APVDGRIGTETMPGVFAQIGTEYGHKYGGTSFELFAKISEKNHAHSTLNP
:. : :
Mb1653c|M.bovis_AF2122/97 NARYRKENSIDDVLASPVVSDPLRLLDICATSDGAAALIVASKSFTEKHL
Rv1627c|M.tuberculosis_H37Rv NARYRKENSIDDVLASPVVSDPLRLLDICATSDGAAALIVASKSFTEKHL
MMAR_2430|M.marinum_M NARYRKEVSVEDVLASQVVAEPLRLLDICATSDGAAALIVASAKFAREHL
MAB_2619|M.abscessus_ATCC_1997 NARFRKESSVADVLASPVVSDPLRLLDICATSDGAAALIVASADFAKKHL
MSMEG_4778|M.smegmatis_MC2_155 NAFRRKEISVEEILASPTLNHPLTQYMFCAPDEGAAAVIMCRADIAHKFT
MAV_0989|M.avium_104 NAFRRKEIPVEEILNSAVLNYPLTQYMFCAPDEGAAAVIMCRADLAHKFT
Mflv_2519|M.gilvum_PYR-GCK NAFRRKEISVEEIMASPVLNYPLRQYMFCAPDEGAAAVIMCRADIAHKYT
Mvan_4139|M.vanbaalenii_PYR-1 NAFRRKEISVEEIMASPVLNYPLRQYMFCAPDEGAAAVIMCRGDIAHRFT
TH_0532|M.thermoresistible__bu NAFRRKEISVEDILNSPVLNYPLTQYMFCAPDEGAAAVIMCRGDIAHRFT
MLBr_02158|M.leprae_Br4923 -------------------TRSLTQCMFCALNKGAFAVIVCLGDIAHKFT
MUL_4955|M.ulcerans_Agy99 LAAYQKRFTLEQIMNDVMIAYPNTRPMCSANCDGAAAAIVCNGETLKSLS
. .* .** * *:. . .
Mb1653c|M.bovis_AF2122/97 GSVAGVPSVRAISTVTPKYPQHLPELPDIATDSTAAVPAPERVFKDQILD
Rv1627c|M.tuberculosis_H37Rv GSVAGVPSVRAISTVTPKYPQHLPELPDIATDSTAAVPAPERVFKDQILD
MMAR_2430|M.marinum_M GSLDGVPSVRAVSTVTPRYPQHLPELPDIATDSTAVVPAPERVFKDQILD
MAB_2619|M.abscessus_ATCC_1997 GSVDGVPSVRAVATITPKYPQHLPELPDIATDSTAVVPAPERVFKDQIVD
MSMEG_4778|M.smegmatis_MC2_155 DKPVYVRASEIRTRTFGAYEVHATSAPPESDAAP-TVYAAR---------
MAV_0989|M.avium_104 DKPVYVRACEIRTRRYGAYEVHATSAPLDEDASP-TVYAAR---------
Mflv_2519|M.gilvum_PYR-GCK DKPVYVRASEIRTRTYGAYEVHATSAPLDEDPSP-TVFAAK---------
Mvan_4139|M.vanbaalenii_PYR-1 DKPVFVRASEIRTRTFGAYEVHATSAPLEEDPSP-TVFAAK---------
TH_0532|M.thermoresistible__bu DKPVYVRASEIRTRRYGAYEVHATSAPLEEDVAP-TVYAAR---------
MLBr_02158|M.leprae_Br4923 DKTVHVHVTEIRTRRYGAYEVHATFAPLVEDASPKRCTIAK---------
MUL_4955|M.ulcerans_Agy99 ARQR--RRAVKVSASVLTTDPYEPGCQVLPNVNTLTRNAAK---------
: : . . ..
Mb1653c|M.bovis_AF2122/97 AAYAEAGIGPEDLSLAEVYDLSTAL-ELDWYEHLGLCPKGEAEALLRSGA
Rv1627c|M.tuberculosis_H37Rv AAYAEAGIGPEDLSLAEVYDLSTAL-ELDWYEHLGLCPKGEAEALLRSGA
MMAR_2430|M.marinum_M AAYTEAGIGPEDLSLAEVYDLSTAL-ELDWYEHLGLCPKGEAEALLRSGA
MAB_2619|M.abscessus_ATCC_1997 AAYAEAGIGPEDISLAEVYDLSTAL-ELDWYEHLGLCAKGEGEQLLRSGA
MSMEG_4778|M.smegmatis_MC2_155 AAYEAAGIGPEDVDIAQLQDTDAGA-EVIHMAETGLCADGDQEKLIAEGA
MAV_0989|M.avium_104 AAYEAAGIGPEDVDVAQLQDTDAGA-EVIHMAETGLCADGEQEKLLADGA
Mflv_2519|M.gilvum_PYR-GCK AAYEAAGIGPEDVDIAQLQDTDAGA-EVIHMAETGLCADGEQEKLLADGA
Mvan_4139|M.vanbaalenii_PYR-1 AAYEAAGIGPEDVDIAQLQDTDAGA-EVIHMAETGLCADGEQEKLLADGA
TH_0532|M.thermoresistible__bu AAYEAAGIGPEDVDIAQLQDTDAGA-EVIHMAETGLCADGEQEKLLAEGA
MLBr_02158|M.leprae_Br4923 AVYRGRQASDLRTSTSPSCRTSVPAPEVIQMDETVLCADGEQEKLASW--
MUL_4955|M.ulcerans_Agy99 TAYEQAGVGPSDLDLVELHDCFATA-ELVHYDNLMLCEQGGAADFFESGA
:.* . . . *: . ** .* :
Mb1653c|M.bovis_AF2122/97 TTLGGRVPVNPSGGLACFGEAIPAQAIAQVCELTWQLRGQATGRQVAD-A
Rv1627c|M.tuberculosis_H37Rv TTLGGRVPVNPSGGLACFGEAIPAQAIAQVCELTWQLRGQATGRQVAD-A
MMAR_2430|M.marinum_M TTIGGRVPVNPSGGLACFGEAIPAQAIAQVCELNWQLRGQATGRQVEN-A
MAB_2619|M.abscessus_ATCC_1997 TTVGGRVPVNASGGLACFGEAIPAQAIAQVCELTWQLRGQAGDRQVEG-A
MSMEG_4778|M.smegmatis_MC2_155 TEIDGSIPVNTDGGLIANGEPIGASGLRQMHELVRQLRGEAGDRQVPGNP
MAV_0989|M.avium_104 TEIHGSMPVNTDGGLIANGEPIGASGLRQVHELVRQLRGEAGERQVPGNP
Mflv_2519|M.gilvum_PYR-GCK TEIHGSIPVNTDGGLIANGEPIGASGLRQVHELVRQLRGEAGDRQVPGAP
Mvan_4139|M.vanbaalenii_PYR-1 TEIHGSIPVNTDGGLIANGEPIGASGLRQVHELVRQLRGQAGDRQVPGQP
TH_0532|M.thermoresistible__bu TEINGSLPINTDGGLIANGEPIGASGLRQIHELVRQLRGQAGDRQVPGNP
MLBr_02158|M.leprae_Br4923 --------------------------------------------------
MUL_4955|M.ulcerans_Agy99 PWRHGSTPVNVSGGLQSKGHPIAATGIANVWEICHQLRGEAGDRQIDN-A
Mb1653c|M.bovis_AF2122/97 KVGVTANQGLFGHGSSVIVAR---
Rv1627c|M.tuberculosis_H37Rv KVGVTANQGLFGHGSSVIVAR---
MMAR_2430|M.marinum_M RVGVTANQGLFGHGSSVIVAR---
MAB_2619|M.abscessus_ATCC_1997 RVGITANQGLFGNGSSVIVAR---
MSMEG_4778|M.smegmatis_MC2_155 RVGLAQVYGAPGTASATILSL---
MAV_0989|M.avium_104 RVGLAQVYGAPGTASATILSL---
Mflv_2519|M.gilvum_PYR-GCK RVGLAQVYGAPGTASATILSL---
Mvan_4139|M.vanbaalenii_PYR-1 RVGLAQVYGAPGTASATILSL---
TH_0532|M.thermoresistible__bu KVGFAQLYGAPGTAAATVLTT---
MLBr_02158|M.leprae_Br4923 ------------------------
MUL_4955|M.ulcerans_Agy99 KVGLAHVIGLGSACGVHILERSAA