For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSDLRVAVLGVGVMGADHVERLSTRIAGVKVAVVNDFIETRAEEIAAGVPGCRVVSDPLEAIADPDVDAV VLATPGPTHDKQLLACLEQRKPVLCEKPLTTDVDSALDVVRREAELGVRLIQVGFMRRFDPEYAELKTLI DAGEFGQPLVLHCVHRNAAVPPSFDSTMIVKDSLVHEVDVTRFLFDEEIASVHILRPTPNPGAPEGIQDP QIAIMKTPSGKHVDVELFVTTGVGYEVRTELVAEKGTAMIGLDVGLIRKGVPGTWGGAIAPDFRVRFGTA YDIEFQRWVAAVRHGIATGSDDYTDGPTAWDGYAAAAVCAAGVESLAGGQPVEVKMVERASINGA
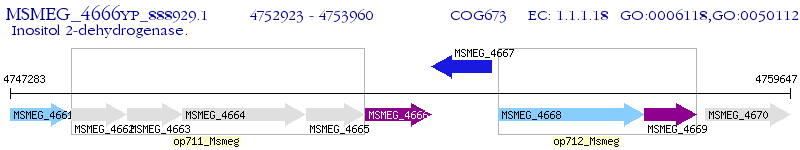
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4666 | - | - | 100% (345) | myo-inositol 2-dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_4650 | - | 2e-21 | 28.57% (343) | oxidoreductase YisS |
| M. smegmatis MC2 155 | MSMEG_6717 | - | 6e-14 | 28.27% (191) | oxidoreductase, Gfo/Idh/MocA family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2598 | - | 1e-137 | 68.33% (341) | inositol 2-dehydrogenase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2306c | - | 5e-11 | 31.25% (144) | putative oxidoreductase |
| M. marinum M | MMAR_2628 | - | 1e-09 | 30.34% (145) | hypothetical protein MMAR_2628 |
| M. avium 104 | MAV_1941 | - | 2e-11 | 27.38% (263) | oxidoreductase family protein, NAD-binding Rossmann fold |
| M. thermoresistible (build 8) | TH_3104 | - | 5e-07 | 28.10% (153) | PUTATIVE oxidoreductase domain protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0387 | - | 1e-146 | 73.04% (345) | inositol 2-dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4666|M.smegmatis_MC2_155 ----MSDLRVAVLGVGVMGADHVERLSTRIAGVKVAVVNDFIETRAEEIA
Mvan_0387|M.vanbaalenii_PYR-1 ----MSEIGVAVLGVGLMGADHVARIASRISGARVAVVNDQVTEKAERLA
Mflv_2598|M.gilvum_PYR-GCK ----MTDLRIAVLGVGVMGADHVARITSRISGARVAVVNDHLVEKAEQLA
MAV_1941|M.avium_104 -----MTLRVGIIGVGWGAHVQVPGFR-AAKGYEPVALCARTPERLERVA
TH_3104|M.thermoresistible__bu -------MRVLVIGTGFGRQAMAPVYR--GLGFDVEVVSPRDESALHRAL
MAB_2306c|M.abscessus_ATCC_199 ---MPESIRWGIVGPGRIAGNVARDFP-LTPRAELAAVASRSLPRAQDFA
MMAR_2628|M.marinum_M MTGHGSPLRIGILGAARIAPQALVKPAQGNVEAVVAAVAARDGARAQAFA
: ::* . .: .
MSMEG_4666|M.smegmatis_MC2_155 AGVPGCRVVSDPLEAIADPDVDAVVLATPGPTHDKQLLACLEQRKPVLCE
Mvan_0387|M.vanbaalenii_PYR-1 ATITGCRAIADPLDAIADRDVDAVVLATPGPTHEKQLLACLEHRKPVLCE
Mflv_2598|M.gilvum_PYR-GCK ASIPGCSAVADPLDAIADADVDAVVLATPGGTHEEQLLACLDQRKPVMCE
MAV_1941|M.avium_104 DKLGIEETSTDWESFVTRDDLDVISVATPTVLHRDMTLAALAAGKPVLCE
TH_3104|M.thermoresistible__bu N-----------------AGVDLVSVHSPPFLHRQHVTAAVDRGYPVLCD
MAB_2306c|M.abscessus_ATCC_199 GAHQIPRAYGSYRELLADPDIDAVYIATPHPQHRQTAIAALTAGKAILVE
MMAR_2628|M.marinum_M AKHGISRVHSSYEALIADPTLDAVYIPLPNGLHGRWTQAALTAGKHVLCE
:* : : * * *.: :: :
MSMEG_4666|M.smegmatis_MC2_155 KPLTTDVDSALDVVRREAELGVRLIQVGFMRRFDPEYAELKTLIDAGEFG
Mvan_0387|M.vanbaalenii_PYR-1 KPLTTDVETSLEVVRREAELGVRLIQVGFMRRFDDEYRALKALIDGGELG
Mflv_2598|M.gilvum_PYR-GCK KPLTTDVSTSLEIARREADLGRPLIQVGFMRRFDDEYVRLKALLDGGELG
MAV_1941|M.avium_104 KPLAGDLDAARDMVRAAAEANVPTACC-FENRWNPDWLAVADRVRSGFLG
TH_3104|M.thermoresistible__bu KPFGRNADEARAMRDHAARRAVPTFLN-FELRHQPARAEFKKLADSGVIG
MAB_2306c|M.abscessus_ATCC_199 KTFTATVSGAQEVIDIARARGVFAMEA-MWTRFQPAIVAAKDLVDDGAIG
MMAR_2628|M.marinum_M KPFTANAAEADEIAQLAATTDRVVMEA-FHYRYHPLTLRIEQILASGELG
*.: : : : * . * :*
MSMEG_4666|M.smegmatis_MC2_155 QPLVLHCVHRN----AAVPPSFDSTMIVKDS-------LVHEVDVTRFLF
Mvan_0387|M.vanbaalenii_PYR-1 NPLVLHCVHRN----PVVPGHFDSAMTVRDS-------LVHEVDVTRFLF
Mflv_2598|M.gilvum_PYR-GCK NPLMMHCVHRN----PGVPAYFDSSLIVKDS-------LVHEVDITRYLF
MAV_1941|M.avium_104 TQYLARVSRSASYWHPSHPLQAHWMYDREQGGGYLAGMLVHDLDFLCSLL
TH_3104|M.thermoresistible__bu TPRHLSWTFFG---AGLRGRPHGWINERDTGGGWIGANGAHLIDYTRYLF
MAB_2306c|M.abscessus_ATCC_199 EVRQVQADLGV----DRPFDREDRLFNPALGGGALLDLGVYVVSLAQYFL
MMAR_2628|M.marinum_M TLQRVETSMCFP-----LPKFSDIRYNYALAGGATMDAGCYAVHMARTFG
. : : :
MSMEG_4666|M.smegmatis_MC2_155 DEEIASVH-ILRP-TPNPGAPEG------IQDPQIAIMKTPSGKHVDVEL
Mvan_0387|M.vanbaalenii_PYR-1 DEEIVSIQ-IVKP-TPNSLAREG------LFDPQIAILRTASGRHVDVEL
Mflv_2598|M.gilvum_PYR-GCK GEEIASVQ-IIKP-TSNPGAPNG------VVDPQIAILRTVSGRHVDVEL
MAV_1941|M.avium_104 GRPVSVCA-EVRTSDPVRELPDGGTLHVTADDTAAVLMRMESGVTAVLSV
TH_3104|M.thermoresistible__bu GSEITGCGGVARIEDPHRPDPQGRPRRATAEDAYSAWFVTENGCTATHDT
MAB_2306c|M.abscessus_ATCC_199 GDPAAVVA-------HGSLFPTG------VDAEAGLLLRYDDGRTATLLC
MMAR_2628|M.marinum_M GGTPEVVS------AQAKRRDSE------VDRAMTAELRFPAGHSGRIQC
. : *
MSMEG_4666|M.smegmatis_MC2_155 FVTTGVGYEVRTELVAEKGTAMIGLDVGLIRKGVPGTWGGAIAPDFRVRF
Mvan_0387|M.vanbaalenii_PYR-1 FVTTGVAYEVRTELVAEKGSVIIGLDVGLVRKSAPGTWGGTITPSFKERF
Mflv_2598|M.gilvum_PYR-GCK FVTTGVAYEVRTEVVGEHGSAIIGLDVGLIRKKGPGSWGGTLTPGFRERF
MAV_1941|M.avium_104 SVMGAHADHYRLELFGSDGTIIGDGDLRSAAYSLGAATDDGLRSLAVDER
TH_3104|M.thermoresistible__bu AFAAAVALPPRMVLTGSAGALELIGDSRLIVHRADADPMTHEFPPAPPR-
MAB_2306c|M.abscessus_ATCC_199 SLLHHTPGQAR--IFGTEGWIDILPRFHHPREIVLHRKGAAPEPMVRPPV
MMAR_2628|M.marinum_M SMWSSHLLNMSARVVGDRGELRVLNPVVPQLFHRLTVQSAIGRRVERFPR
. : . *
MSMEG_4666|M.smegmatis_MC2_155 GTAYDIEFQRWVAAVRHGIATGSDDYTDGPTAWDGYAAAAVCAAGVESLA
Mvan_0387|M.vanbaalenii_PYR-1 GQAYDTEIQRWVDAVRSGGTTG--IYTDGPTAWDGYAATAVCEAGVQALQ
Mflv_2598|M.gilvum_PYR-GCK GPAYDTEIQRWVDAVHSG------TNVCGPTAWDGYAAAAVCAAGVESLE
MAV_1941|M.avium_104 EPAHPDRLPRGLAGHASR--------AMALMLQDWLPAFDGAPSNVATFE
TH_3104|M.thermoresistible__bu -SFYEPALTPWLEQVADA-------------LRTGRPATPSFDDGVAVAE
MAB_2306c|M.abscessus_ATCC_199 GGGYSHELAEVTECLLNG-----------RAQSSVMPLADTLAVQRVLNT
MMAR_2628|M.marinum_M RASYAYQLDAFTGAILRG--------------EAVKTTPQDAVQNMTVID
: : .
MSMEG_4666|M.smegmatis_MC2_155 GGQPVEVKMVERASINGA----------
Mvan_0387|M.vanbaalenii_PYR-1 SGQPVAVSMVDRVSIPGA----------
Mflv_2598|M.gilvum_PYR-GCK TGLPVDVQLADEVTSPSPQGVRESDGCR
MAV_1941|M.avium_104 DG-LLSLAIIDAARRSAEGGGWEPVQA-
TH_3104|M.thermoresistible__bu VMDRLRANVIEQVMQ-------------
MAB_2306c|M.abscessus_ATCC_199 ACEQLGVDHTEDPADLD-----------
MMAR_2628|M.marinum_M AIYRAAGLPLRNPS--------------