For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PESIRWGIVGPGRIAGNVARDFPLTPRAELAAVASRSLPRAQDFAGAHQIPRAYGSYRELLADPDIDAVY IATPHPQHRQTAIAALTAGKAILVEKTFTATVSGAQEVIDIARARGVFAMEAMWTRFQPAIVAAKDLVDD GAIGEVRQVQADLGVDRPFDREDRLFNPALGGGALLDLGVYVVSLAQYFLGDPAAVVAHGSLFPTGVDAE AGLLLRYDDGRTATLLCSLLHHTPGQARIFGTEGWIDILPRFHHPREIVLHRKGAAPEPMVRPPVGGGYS HELAEVTECLLNGRAQSSVMPLADTLAVQRVLNTACEQLGVDHTEDPADLD*
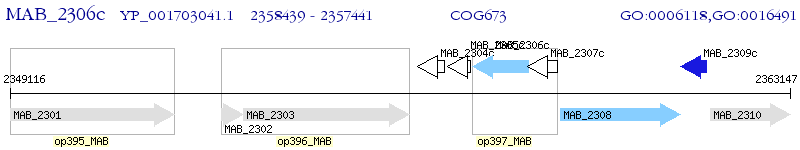
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2306c | - | - | 100% (332) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_2612 | - | 2e-10 | 25.56% (223) | putative oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2611 | - | 6e-25 | 32.46% (228) | oxidoreductase domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_2628 | - | 1e-33 | 33.33% (300) | hypothetical protein MMAR_2628 |
| M. avium 104 | MAV_2987 | - | 2e-32 | 33.44% (314) | NAD binding oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_4652 | - | 5e-22 | 30.64% (235) | D-oliose 4-ketoreductase |
| M. thermoresistible (build 8) | TH_3104 | - | 2e-05 | 27.56% (127) | PUTATIVE oxidoreductase domain protein |
| M. ulcerans Agy99 | MUL_3103 | - | 4e-05 | 28.57% (140) | hypothetical protein MUL_3103 |
| M. vanbaalenii PYR-1 | Mvan_3970 | - | 6e-26 | 28.40% (324) | oxidoreductase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2611|M.gilvum_PYR-GCK --------MT-LRIGVLGASRIAEAAIVEP-ARELGFRLVAVAARDPQRA
Mvan_3970|M.vanbaalenii_PYR-1 --------MS-LRIGILGASRIAASAIVEP-ARELGHRLIAVAARDPLRT
MSMEG_4652|M.smegmatis_MC2_155 --------MTGLRIGILGASRIAEKAIVGP-AEDLGHRLVAVAARDRRRA
MMAR_2628|M.marinum_M ----MTGHGSPLRIGILGAARIAPQALVKPAQGNVEAVVAAVAARDGARA
MUL_3103|M.ulcerans_Agy99 --------------------------------------------------
MAV_2987|M.avium_104 MSAMAEVHAPKLRIGILGAARIAPLALIKPSAENPDVVVAAVAARDASRA
MAB_2306c|M.abscessus_ATCC_199 -------MPESIRWGIVGPGRIAG-NVARDFPLTPRAELAAVASRSLPRA
TH_3104|M.thermoresistible__bu -----------MRVLVIGTG--FGRQAMAPVYRGLGFDVEVVSPRD----
Mflv_2611|M.gilvum_PYR-GCK NIFADTYGVERVVPTYQDVVTDPEVDVVYNPLANSLHAPWNLAAVAAGKP
Mvan_3970|M.vanbaalenii_PYR-1 QAFADKYGVERVLASYQDVVTDPQVDIVYNPLANALHAPWNLAAVAAGKP
MSMEG_4652|M.smegmatis_MC2_155 EAFAEKYGVQRVLDSYRDVVEDPDVDMVYNPLANGLHAPWNLAAIAAGKP
MMAR_2628|M.marinum_M QAFAAKHGISRVHSSYEALIADPTLDAVYIPLPNGLHGRWTQAALTAGKH
MUL_3103|M.ulcerans_Agy99 --------------------------------------------------
MAV_2987|M.avium_104 RAFATRHGIATVHDHYEALIADPDIDAIYNPLPNGLHGRWTRAALAAGKH
MAB_2306c|M.abscessus_ATCC_199 QDFAGAHQIPRAYGSYRELLADPDIDAVYIATPHPQHRQTAIAALTAGKA
TH_3104|M.thermoresistible__bu -----ESALHRALN--------AGVDLVSVHSPPFLHRQHVTAAVDRGYP
Mflv_2611|M.gilvum_PYR-GCK VLSEKPFARNRAEAARVASAADAAGVTVMEGFHYLFHPVTQRALALAGDG
Mvan_3970|M.vanbaalenii_PYR-1 VLTEKPYARDRGEAARVAAAADAAGVTVMEGFHYLFHPVTQRALSLAGDG
MSMEG_4652|M.smegmatis_MC2_155 VLSEKPFARNESEAATVAQAARDAGVPVLEGFHYHFHPVTQRAFAVAAGG
MMAR_2628|M.marinum_M VLCEKPFTANAAEADEIAQLAATTDRVVMEAFHYRYHPLTLRIEQILASG
MUL_3103|M.ulcerans_Agy99 --------------------------------------------------
MAV_2987|M.avium_104 VLCEKPFTANAAEAREIADLAEHSDRVVMEAFHYRYHPLTLRAEEIIASG
MAB_2306c|M.abscessus_ATCC_199 ILVEKTFTATVSGAQEVIDIARARGVFAMEAMWTRFQPAIVAAKDLVDDG
TH_3104|M.thermoresistible__bu VLCDKPFGRNADEARAMRDHAARRAVPTFLNFELRHQPARAEFKKLADSG
Mflv_2611|M.gilvum_PYR-GCK TLGELRRVEVRMGMPEPKP--DDPRWSLELAGGALMDLGCYGVHVLRRFG
Mvan_3970|M.vanbaalenii_PYR-1 TLGELTRIEVRMGMPAPQS--DDPRWSLELAGGALMDLGCYGLHVMRRFG
MSMEG_4652|M.smegmatis_MC2_155 ELGEITRVEVRMAMPSPAD--DDPRWSLDLAGGAVMDLGCYGIHVMRALG
MMAR_2628|M.marinum_M ELGTLQRVETSMCFPLPKF--SDIRYNYALAGGATMDAGCYAVHMARTFG
MUL_3103|M.ulcerans_Agy99 -----------MRFPLPKF--SDIRYNYALAGGATMDAGCYAVHMARTFG
MAV_2987|M.avium_104 ELGTLQHVEAALCFPLPKF--SDIRYDYALAGGATMDAGCYAVHMARTFG
MAB_2306c|M.abscessus_ATCC_199 AIGEVRQVQADLGVDRPFDR-EDRLFNPALGGGALLDLGVYVVSLAQYFL
TH_3104|M.thermoresistible__bu VIGTPRHLSWTFFGAGLRGRPHGWINERDTGGGWIGANGAHLIDYTRYLF
: . . .** * : : : :
Mflv_2611|M.gilvum_PYR-GCK --------------VPSILSATAVQRSAGVDATLDAEVVFPSGLTGLTAN
Mvan_3970|M.vanbaalenii_PYR-1 --------------TPSVLAATAVQRTPGVDERFDAELVFPSGLTGLTTN
MSMEG_4652|M.smegmatis_MC2_155 AMRVDGL-----GGEPKVLGARAEERTPGVDAWCEVDFVFPEGGTGVSTN
MMAR_2628|M.marinum_M ------------GGTPEVVSAQAKRRDSEVDRAMTAELRFPAGHSGRIQC
MUL_3103|M.ulcerans_Agy99 ------------GGTPEVVSAQAKRRDSEVDRAMTAELRFPAGHSGRIEF
MAV_2987|M.avium_104 ------------GRSPEVVSAQAKLRGPKVDRAMTAELRFPAGHTGRVRC
MAB_2306c|M.abscessus_ATCC_199 G-------------DPAAVVAHGSLFPTGVDAEAGLLLRYDDGRTATLLC
TH_3104|M.thermoresistible__bu GSEITGCGGVARIEDPHRPDPQGRPRRATAEDAYSAWFVTENGCTATHDT
* . . . .: . * :.
Mflv_2611|M.gilvum_PYR-GCK SMVQDDFS-FTLRLIGSRGEAFVHN--FIKPRDDDRLTVQTDDGTRVERF
Mvan_3970|M.vanbaalenii_PYR-1 SMVDDEYS-FTLRLIGTRGEAFVHD--FIKPHDDDRLTLHTEDGTTVERH
MSMEG_4652|M.smegmatis_MC2_155 SMVADDYS-FTITITGSDGEAVVHD--FIRPNLDDRLTVMTGSGVRVEHL
MMAR_2628|M.marinum_M SMWSSHLLNMSARVVGDRGELRVLN--PVVPQLFHRLTVQSAIGRRVERF
MUL_3103|M.ulcerans_Agy99 SMWSSHLLNMSARVVGDRGELRVLN--PVVPHLFHRLTVQSAIGRRVERF
MAV_2987|M.avium_104 SMWSTRLLQISARVVGEHGELRIVN--PVLPQAFHRLTVRSRDGRRTERF
MAB_2306c|M.abscessus_ATCC_199 SLLHHTPG--QARIFGTEGWIDILP--RFHHPREIVLHRKGAAPEPMVRP
TH_3104|M.thermoresistible__bu AFAAAVALPPRMVLTGSAGALELIGDSRLIVHRADADPMTHEFPPAPPRS
:: : * * : . :
Mflv_2611|M.gilvum_PYR-GCK GTRASYTYQLEFFARHVLHG--APLPLDTTDAVENMAVIDEIYRAAGLPV
Mvan_3970|M.vanbaalenii_PYR-1 GTRTSYTYQLEAFTDHVERG--TPIPLDTADAVANMALIDEAYRAAGMSP
MSMEG_4652|M.smegmatis_MC2_155 GTRPSYTYQLEAFAAHVRDG--APLPFGVDDAVTNMAFVDKAYRAAGLAP
MMAR_2628|M.marinum_M PRRASYAYQLDAFTGAILRG--EAVKTTPQDAVQNMTVIDAIYRAAGLPL
MUL_3103|M.ulcerans_Agy99 PRRASYVYQLDAFTGAILRG--EAVKTTPQDAVQNMTVIDAIYRAAGLPL
MAV_2987|M.avium_104 ERRASYAYQLDAFAAGVLRG--EPVKTSARDAVDNMTVIDAIYRAAGLPL
MAB_2306c|M.abscessus_ATCC_199 PVGGGYSHELAEVTECLLNGRAQSSVMPLADTLAVQRVLNTACEQLGVDH
TH_3104|M.thermoresistible__bu FYEPALTPWLEQVADALRTG--RPATPSFDDGVAVAEVMDRLRANVIEQV
. * .: : * . * : .::
Mflv_2611|M.gilvum_PYR-GCK R-------
Mvan_3970|M.vanbaalenii_PYR-1 R-------
MSMEG_4652|M.smegmatis_MC2_155 R-------
MMAR_2628|M.marinum_M RNPS----
MUL_3103|M.ulcerans_Agy99 RNPS----
MAV_2987|M.avium_104 RYPT----
MAB_2306c|M.abscessus_ATCC_199 TEDPADLD
TH_3104|M.thermoresistible__bu MQ------