For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MWRQPKGVWAVAFASVVAFMGIGLVDPILKPIADNLDATPSQVSLLFTSYMAVMGVAMLITGVVSSRIGP KRTLLVGLVIIIAGAGLAGMSDSVMEIVGWRALWGLGNALFIATALATIVNSAKGSVAQAIILYEAALGL GIAVGPLVGGVLGSISWRGPFFGVSALMALALVVTAFLLPETPRPQHATSLADPFRALRHRGLFGVGLTA LLYNFGFFTLLAFTPFPLDMTAHQIGLIFFGWGVALAFTSVVVAPRLQHRFGTLPTLLVNLAVMSATLVV MALGTDNKAVLAASVVVAGLWCGINNTLITETVMKAAPVERGVASAAYSFMRFCGAAVAPWLAGVLGEEI SVHLPYWVGAAAVAAGAAVLFAVRSHLAGVDAEESELDELTDEATAVTVGND
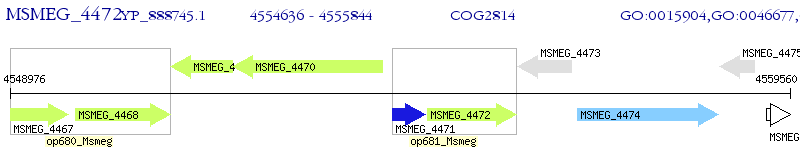
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4472 | - | - | 100% (402) | multidrug-efflux transporter protein |
| M. smegmatis MC2 155 | MSMEG_2991 | - | 4e-27 | 26.96% (408) | permease of the major facilitator superfamily protein |
| M. smegmatis MC2 155 | MSMEG_2137 | - | 1e-18 | 23.00% (387) | permease of the major facilitator superfamily protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3755 | - | 5e-13 | 26.55% (177) | hypothetical protein Mb3755 |
| M. gilvum PYR-GCK | Mflv_2728 | - | 0.0 | 85.71% (399) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv3728 | - | 6e-13 | 26.55% (177) | hypothetical protein Rv3728 |
| M. leprae Br4923 | MLBr_01562 | - | 8e-12 | 30.00% (150) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | MAB_3449c | - | 4e-24 | 26.02% (392) | putative transporter |
| M. marinum M | MMAR_1146 | - | 3e-20 | 26.24% (423) | integral membrane transport protein |
| M. avium 104 | MAV_3974 | - | 2e-15 | 25.88% (371) | permease of the major facilitator superfamily protein |
| M. thermoresistible (build 8) | TH_3412 | - | 7e-16 | 23.92% (464) | PUTATIVE putative major facilitator superfamily MFS_1 |
| M. ulcerans Agy99 | MUL_0911 | - | 3e-20 | 26.82% (425) | integral membrane transport protein |
| M. vanbaalenii PYR-1 | Mvan_5021 | - | 7e-20 | 23.82% (424) | EmrB/QacA family drug resistance transporter |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4472|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2728|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1146|M.marinum_M --------------------------------------------MRIQPV
MUL_0911|M.ulcerans_Agy99 --------------------------------------------MRIQPV
TH_3412|M.thermoresistible__bu -------------------------------------------MTQCAVR
Mb3755|M.bovis_AF2122/97 ----------------MHTVATNNAAPVIAAGPVGPSRRRRRVHAPLTRR
Rv3728|M.tuberculosis_H37Rv ----------------MHTVATNNAAPVIAAGPVGPSRRRRRVHAPLTRR
MAB_3449c|M.abscessus_ATCC_199 -------------------------------------MSSCREGNADNAT
MAV_3974|M.avium_104 --------------------------------------------------
Mvan_5021|M.vanbaalenii_PYR-1 -------------------------------------------------M
MLBr_01562|M.leprae_Br4923 MVTLYDHERAVHNWTAARSDRPAPSRLTQQVEPASERTSKYPTLLPSGRF
MSMEG_4472|M.smegmatis_MC2_155 MWR-QPKGVWAVAFASVVAFMGIGLVDPILKPIADNLDATPSQVSLLFTS
Mflv_2728|M.gilvum_PYR-GCK MLRSQPKAVWAVAFASVVAFMGIGLVDPILKPIADNLDASPSQVSLLFTS
MMAR_1146|M.marinum_M FRDHPIPALAVICLSVFVISVDATIVNVALPTLSRQLGADTAQLQWIVDA
MUL_0911|M.ulcerans_Agy99 FRDHPILALAVICLSVFVISVDATIVNVALPTLSRELGADTAQLQWIVDA
TH_3412|M.thermoresistible__bu AGAREWAGLGVLAVPTVLLGLDVTVLYLVLPAVAADLAPSPVQTLWIMDA
Mb3755|M.bovis_AF2122/97 RQPSSSAVLLVAAFGAFLAFLDSTIVNVAFPDIQRHFHSDISDLSWMLNA
Rv3728|M.tuberculosis_H37Rv RQPSSSAVLLVAAFGAFLAFLDSTIVNVAFPDIQRHFHSDISDLSWMLNA
MAB_3449c|M.abscessus_ATCC_199 VKPRLPWEIWVLVVASLVIALGFGVVAPALPQYARSFGVSVTAATAVVSS
MAV_3974|M.avium_104 -----------------MVALGYGVISPALPTFARSFGVSIKAVTFLVTV
Mvan_5021|M.vanbaalenii_PYR-1 TPRRKAIILVSCCLSLLIVSMDATIVNVAIPSIRDDLSATPAQMQWVVDV
MLBr_01562|M.leprae_Br4923 PSGRFIAAVIVIGGMQLLATMDSTVAIVALPKIQNELSLSDAGRSWGITA
: :. : : : .
MSMEG_4472|M.smegmatis_MC2_155 YMAVMGVAMLITGVVSSRIGPKRTLLVGLVIIIAGAGLAGMSDSVMEIVG
Mflv_2728|M.gilvum_PYR-GCK YMAVMGVAMLITGVVSSRIGPKRTLLLGLVIIIAGAGLAGTSDSVMGIVG
MMAR_1146|M.marinum_M YTLVMSGLLLSAGSLSDRYGRRGWLSAGLILFALTSALAAQVNSADTLIA
MUL_0911|M.ulcerans_Agy99 YTLVMSGLLLSAGSLSDRYGRRGWLSAGLILFALTSALAAQVNSADTLVA
TH_3412|M.thermoresistible__bu YGFFIAGFLITMGTLGDRIGRRRLLTIGVTAFAGASVLAALAPSAELLIV
Mb3755|M.bovis_AF2122/97 YNIVFAAFLVAAGRLADLMGRKRVFILGVALFTVASGLCAIAESVGELVA
Rv3728|M.tuberculosis_H37Rv YNIVFAAFLVAAGRLADLMGRKRVFILGVALFTVASGLCAIAESVGELVA
MAB_3449c|M.abscessus_ATCC_199 FAAFRLVFAPVAGALVQRLGERWVYMSGLLIVAASTGICAFVHSYWQLLV
MAV_3974|M.avium_104 FSLSRLCFAPISGLLTERLGERRIYIGGLLIVAVSTAACAFSQAYWQLML
Mvan_5021|M.vanbaalenii_PYR-1 YTLVLASLLMLSGATGDRFGRRRVFQIGLAVFALGSLACSLAPTIDALIG
MLBr_01562|M.leprae_Br4923 YVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCAVAWDEVTLVI
: * . * : *: . : .. ::
MSMEG_4472|M.smegmatis_MC2_155 WRALWGLGNALFIATALATIVNSAK--GSVAQAIILYEAALGLGIAVGPL
Mflv_2728|M.gilvum_PYR-GCK WRALWGLGNALFIATALATIVSSAK--GSVAQAIILYEAALGVGIAVGPL
MMAR_1146|M.marinum_M ARAAMGVGAAVIFPTTLGLITNIFTDPVPRAKAIGLWTAMVGVGVAVGPI
MUL_0911|M.ulcerans_Agy99 ARAAMGVGAAVIFPTTLGLITNIFTDPVPRAKAIGLWTAMVGVGVAVGPI
TH_3412|M.thermoresistible__bu ARALLGVAGATLMPSTLSLISNMFVDAAQRALAIGVWATMFAVGMAAGPV
Mb3755|M.bovis_AF2122/97 FRVLQGIGAAVLVPASLGLVVEAFP-AERRAHGVNLWGAAGAIAAGLGPP
Rv3728|M.tuberculosis_H37Rv FRVLQGIGAAVLVPASLGLVVEAFP-AERRAHGVNLWGAAGAIAAGLGPP
MAB_3449c|M.abscessus_ATCC_199 FRSVGGVGSTMFTVSAAALLVRIAP-EEIRGRAQGLYGSSFLLGMVAGPA
MAV_3974|M.avium_104 CRVFSGVGSTMFYVSALGLMIHISP-ADARGRIAGLFTTSFMVGAVGGPA
Mvan_5021|M.vanbaalenii_PYR-1 ARLLQGIGGSMLNPVALSIISQIFTQPVERARALGIWGGVTGISMAAGPI
MLBr_01562|M.leprae_Br4923 ARLLQGIGSAIASPTGLALVATTFSKGSARNTATAVFAAMTAIGSVMGLV
* *:. : . : :: :. *
MSMEG_4472|M.smegmatis_MC2_155 VGGVLGSISWRGPFFGVSALMALALVVTAFLLPETPRPQ-----------
Mflv_2728|M.gilvum_PYR-GCK VGGVLGSVSWRGPFFGVSVLMVVALVVTAFLLPATPPAA-----------
MMAR_1146|M.marinum_M TGGWLLEHFSWGSIFLVNVPIAVAAMAGAILFVPTSRDPAAPRV-DVPGL
MUL_0911|M.ulcerans_Agy99 TGGWLLEHFSWGSIFLVNVPIAVAAMAGAILFVPTSRDPAAPRV-DVPGL
TH_3412|M.thermoresistible__bu VGGALVDRFWWGAAFLLAVPVAAAVLLGARLLP-EYADPQAGRL-DLRSV
Mb3755|M.bovis_AF2122/97 IGGALIEADGWRWVFLVNLPLGVFAVLAARRALVENRAAGRRRVPDVRGA
Rv3728|M.tuberculosis_H37Rv IGGALIEADGWRWVFLVNLPLGVFAVLAARRALVENRAAGRRRVPDVRGA
MAB_3449c|M.abscessus_ATCC_199 LGSAVVGLSLSAPFIIYAAALVIVVMLVYFGLRRSTLLDVADEHHGTPVR
MAV_3974|M.avium_104 VGGLAAGWGLTAPFVVYGVAMLGVALVLFLGLRNSALAAPPPPTRST-VT
Mvan_5021|M.vanbaalenii_PYR-1 VGGLLIDTIGWRSVFWINLPICAAAILLTAIFVPESKSQTMRNVDPIG-Q
MLBr_01562|M.leprae_Br4923 VGGALTEVS-WRLAFLVNVPIGLVMIYLARTALHETHKERMKLD--AAGA
*. . : :
MSMEG_4472|M.smegmatis_MC2_155 --HATSLADPFRALRHRGLFGVG------------------------LTA
Mflv_2728|M.gilvum_PYR-GCK --RATTLADPFRALRHRGLFGVG------------------------VTA
MMAR_1146|M.marinum_M MLSAVGITALVYTIIEAPNWGWTSTRASGGFIAAAIVLVGFALWERRSSH
MUL_0911|M.ulcerans_Agy99 MLSAVGITALVYTIIEAPNWGWTSTRASGGFIAAAIVLVGFALWERRSSH
TH_3412|M.thermoresistible__bu ALSLLAILAAIYAVKHVAVHGVDGVAVAACALAAAAATV-FVHRQRRLDA
Mb3755|M.bovis_AF2122/97 VLLAFALGLLTLGLIKGPDWGWASLPTSGSLLAAAVAMVGFVMSSRHHPA
Rv3728|M.tuberculosis_H37Rv VLLAFALGLLTLGLIKGPDWGWASLPTSGSLLAAAVAMVGFVMSSRHHPA
MAB_3449c|M.abscessus_ATCC_199 LTSALRHRTFLAALMSNFSAGWA---------------------------
MAV_3974|M.avium_104 MREALRVRAYQSALLSNFATGWS---------------------------
Mvan_5021|M.vanbaalenii_PYR-1 GLAIMFLFGTVFTLIEGPVLGWTNPRVVVAAVVVALALVAFLRFESRRHD
MLBr_01562|M.leprae_Br4923 ILATLACTAAVFAFSMGPEKGWISLTTIGSGLVALVAAIAFAVVERTAEN
. *
MSMEG_4472|M.smegmatis_MC2_155 LLYNFGFFTLLAFTPFPLDMTAHQIGLIFFGWGVALAFTSVVVAPRLQHR
Mflv_2728|M.gilvum_PYR-GCK LLYNFGFFTLLAFTPFPLDMGAQQIGLIFFGWGVALAVTSVFVAPRLQRR
MMAR_1146|M.marinum_M PMLDVSVFANRRFSGGSLAVTAGFLTLFGFIFVITQYFQFIKDYTALQTG
MUL_0911|M.ulcerans_Agy99 PMLDVSVFANRRFSGGSLAVTAGFLTLFGFIFVITQYFQFIKDYTALQTG
TH_3412|M.thermoresistible__bu PLLDVTLFADRAFTAALTVLLIGLIGVGGTMYLVTQFLQLVEGLTPVRAG
Mb3755|M.bovis_AF2122/97 PMVEPTLLRIQSFVAGTGLTAVASAGFYAYLLTHVLFLNYVWGYTLLEAG
Rv3728|M.tuberculosis_H37Rv PMVEPTLLRIQSFVAGTGLTAVASAGFYAYLLTHVLFLNYVWGYTLLEAG
MAB_3449c|M.abscessus_ATCC_199 -IFGIRGALLPLLVVEALHQRPGAAGLVFAAFAAGDVSTAFLAG-SWSDD
MAV_3974|M.avium_104 -AFGLRMALVPLFVSDVMGRGIGTIGVVLAAFAGGNALAVVPSG-YLSDR
Mvan_5021|M.vanbaalenii_PYR-1 PFLDLRFFRSIPFTTATVIAISAFAAWGAFLFLMSLYLQGERGFSAMHTG
MLBr_01562|M.leprae_Br4923 PVVPFDLFRDRNRLVTFIAIFLAGGVIFSLTVSIGLYIQDILGYSALRAG
MSMEG_4472|M.smegmatis_MC2_155 FGTLP-TLLVNLAVMSATLVVMALGTDNKAVLAASVVVAGLWCGINNT--
Mflv_2728|M.gilvum_PYR-GCK FGTVP-TLLVNLIAMSATLAVMAVGADHKAVLAACVVVAGLWIGINNT--
MMAR_1146|M.marinum_M VRLLPVALSIALAAILGPRLVERVGTTAVVAAGLAVFAAALAWASTAD-A
MUL_0911|M.ulcerans_Agy99 VRLLPVAFSIALAAILGPRLVERVGTTAVVAAGLAVFAAALAWASTAD-A
TH_3412|M.thermoresistible__bu WWMGPPALAMFVAAIGAPLIARRVRPGVVMAATLGVSVLGYAVLASAGRG
Mb3755|M.bovis_AF2122/97 MAVAPAALVAAVVAAVLGRVADRHGYRFIVGIGALIWAASLLWYLKVVGS
Rv3728|M.tuberculosis_H37Rv MAVAPAALVAAVVAAVLGRVADRHGYRFIVGIGALIWAASLLWYLKVVGS
MAB_3449c|M.abscessus_ATCC_199 IGRKPLVVAGLVVCGSTVVAMGFVSSLPVLVVLSLIGGIGAGLYASPQ--
MAV_3974|M.avium_104 MGRRTLLIVGLVTSGAATVWLGFVASLPVFLVAAGVVGVVTGIYMSPL--
Mvan_5021|M.vanbaalenii_PYR-1 LIYLPIAIGALLFSPLSGRLVGRYGARPSLVIAGVTITAAATMLTFLTAT
MLBr_01562|M.leprae_Br4923 IGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLLMVAMLYGWAFMNR
. . :
MSMEG_4472|M.smegmatis_MC2_155 -------------------------LITETVMKAAPVER-GVASAAYSFM
Mflv_2728|M.gilvum_PYR-GCK -------------------------LITEAVMKAAPVER-GVASAAYSFM
MMAR_1146|M.marinum_M TTPYTQIALQMLLLGGG--LGLTTAPATEAIMGSLPADRAGVGSAVNDTT
MUL_0911|M.ulcerans_Agy99 TTPYTQITLQMLLLGGG--LGLTTAPATEAIMGSLPADRAGVGSAVNDTT
TH_3412|M.thermoresistible__bu DGPAVVVAFAFVYLG----LGAIAALGTDIVVGAAPAAKSGSAAAMSETV
Mb3755|M.bovis_AF2122/97 QPDFLGEWLPGQILQGIGVGATFPLLGSAALARLAKGGSYATASAVTGTI
Rv3728|M.tuberculosis_H37Rv QPDFLGEWLPGQILQGIGVGATFPLLGSAALARLAKGGSYATASAVTGTI
MAB_3449c|M.abscessus_ATCC_199 --------------------------QAAVADVVGSRGRAGTALATFQMT
MAV_3974|M.avium_104 --------------------------QAAVADILGNEARAGLPVATVQMM
Mvan_5021|M.vanbaalenii_PYR-1 TPVWSLLMVFAVFGIG---FSMVNAPVTNAAVSGMPLDRAGAASAVTSTS
MLBr_01562|M.leprae_Br4923 GVPYFPNLLAPIVVGGIG-IGMAVVPLTLSAIAGVGFDRIGPVSAIALML
: . *
MSMEG_4472|M.smegmatis_MC2_155 RFCGAAVAPWLAGVLG----------------------------------
Mflv_2728|M.gilvum_PYR-GCK RFGGAAVAPWLAGVLG----------------------------------
MMAR_1146|M.marinum_M RELGGTLGVAIAGSVFASVYS----------------------------G
MUL_0911|M.ulcerans_Agy99 RELGGTLGVAIAGSVFASVYS----------------------------G
TH_3412|M.thermoresistible__bu QELGIAVGVALLGSVTAAVYR----------------------------S
Mb3755|M.bovis_AF2122/97 RQVGAVIGVAVLVILVGTPAPGAAEEALRHRWALAAICFVAVGIGALSLG
Rv3728|M.tuberculosis_H37Rv RQVGAVIGVAVLVILVGTPAPGAAEEALRHGWALAAICFVAVGIGALSLG
MAB_3449c|M.abscessus_ATCC_199 SDIGLVVGPVVAGAIAEHTS------------------------------
MAV_3974|M.avium_104 SDLGAIVGSMAVGWAAEQIG------------------------------
Mvan_5021|M.vanbaalenii_PYR-1 RQVGVSIGVALCGSLAGSAMATGG------------------------AD
MLBr_01562|M.leprae_Br4923 QSLGGPLVLAVIQAVITSRTLYLG-------------------------G
* :
MSMEG_4472|M.smegmatis_MC2_155 --EEISVHLPYWVGAAAVAAGAAVLFAVRSHLAGVDAEESELDELTDEAT
Mflv_2728|M.gilvum_PYR-GCK --ETFSVHVPFWVGAGAVLLAAGALAATRTHLRHLDDEETGIDELTDEAR
MMAR_1146|M.marinum_M QLGSASTLAT-LPSDVRSAMKGSMAVAHKVIDQLPAGPAAGVRDAVNHAF
MUL_0911|M.ulcerans_Agy99 QLGSASALAT-LPSDVRSAMKGSMAVAHKVIDQLPAGPAAGVRDAVNHAF
TH_3412|M.thermoresistible__bu RMTGPEGVAEGVGGPDAERLAESLPAAVSVADRVPGEAVR----LAQDAF
Mb3755|M.bovis_AF2122/97 RIRPVPAAVEPPPGPPVAPLGARRPPRPAPVASPAAAVAPTPKTSREVNL
Rv3728|M.tuberculosis_H37Rv RIRPVPAAVEPPPGPPVAPLGARRPPRPAPVASPAAAVAPTPKTSREVNL
MAB_3449c|M.abscessus_ATCC_199 -----YGWAFVVGGAMPLIAAVAWVFAPETLGRLASPQVRSMQEVAEDVG
MAV_3974|M.avium_104 -----YGWGFFISGVVLLIAAVGWVMAPET--RTATELEADLMAASRTSS
Mvan_5021|M.vanbaalenii_PYR-1 FAAAARPLWFVCMALGLVILGLGLFATSQRALRSAERLAPLVSGAPEPVG
MLBr_01562|M.leprae_Br4923 TTGPVKFMNDAQLRALDHGYTYGLLWVAGAVVIVGVAALFIGYTPEQVAY
MSMEG_4472|M.smegmatis_MC2_155 AVTVGND-------------------------------------------
Mflv_2728|M.gilvum_PYR-GCK ALTVGSDS------------------------------------------
MMAR_1146|M.marinum_M LDGLQVAT-------LVCAGIALAAAIVVGWLLPARAGQPTPAQTEPALA
MUL_0911|M.ulcerans_Agy99 LDGLQVAT-------LVCAGIALAAAIVVGWLLPARAGQPTPAQTEPALA
TH_3412|M.thermoresistible__bu AAGVNIAA-------LIGG----AAVVVVAVVCATALRHLRPLD------
Mb3755|M.bovis_AF2122/97 LEALRFARPDTQQIELQAGSYLFHAGDVSDALYVVRSGRLQVLAGDGAKD
Rv3728|M.tuberculosis_H37Rv LEALRFARPDTQQIELQAGSYLFHAGDVSDALYVVRSGRLQVLAGDGAKD
MAB_3449c|M.abscessus_ATCC_199 GPER----------------------------------------------
MAV_3974|M.avium_104 RCEQQL--------------------------------------------
Mvan_5021|M.vanbaalenii_PYR-1 GAHVR---------------------------------------------
MLBr_01562|M.leprae_Br4923 AQEVKEAVDAGEL-------------------------------------
MSMEG_4472|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2728|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1146|M.marinum_M QA------------------------------------------------
MUL_0911|M.ulcerans_Agy99 QV------------------------------------------------
TH_3412|M.thermoresistible__bu --------------------------------------------------
Mb3755|M.bovis_AF2122/97 EVVAELGRGQVVGELGVLLDAPRSASVRAVRDSSLMRVTKAEFAKIADAG
Rv3728|M.tuberculosis_H37Rv EVVAELGRGQVVGELGVLLDAPRSASVRAVRDSSLMRVTKAEFAKIADAG
MAB_3449c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3974|M.avium_104 --------------------------------------------------
Mvan_5021|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
MSMEG_4472|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2728|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1146|M.marinum_M --------------------------------------------------
MUL_0911|M.ulcerans_Agy99 --------------------------------------------------
TH_3412|M.thermoresistible__bu --------------------------------------------------
Mb3755|M.bovis_AF2122/97 VLGALAGVLAKRQHQTRVASQRTTPEVVVAVVGVDANAPVAMVATELCRA
Rv3728|M.tuberculosis_H37Rv VLGALAGVLAKRQHQTRVASQRTTPEVVVAVVGVDANAPVAMVATELCRA
MAB_3449c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3974|M.avium_104 --------------------------------------------------
Mvan_5021|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
MSMEG_4472|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2728|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1146|M.marinum_M --------------------------------------------------
MUL_0911|M.ulcerans_Agy99 --------------------------------------------------
TH_3412|M.thermoresistible__bu --------------------------------------------------
Mb3755|M.bovis_AF2122/97 LSTRLRAVAPGRVDCDGLERAEQTADRVVLHAAVGDARWREFCLRVADRV
Rv3728|M.tuberculosis_H37Rv LSTRLRAVAPGRVDCDGLERAEQTADRVVLHAAVGDARWREFCLRVADRV
MAB_3449c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3974|M.avium_104 --------------------------------------------------
Mvan_5021|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
MSMEG_4472|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2728|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1146|M.marinum_M --------------------------------------------------
MUL_0911|M.ulcerans_Agy99 --------------------------------------------------
TH_3412|M.thermoresistible__bu --------------------------------------------------
Mb3755|M.bovis_AF2122/97 VLVASNPAVPVAPLPTRATGADLVLAGRPAGREHRRAWEQLITPRSMHVV
Rv3728|M.tuberculosis_H37Rv VLVASNPAVPVAPLPTRATGADLVLAGRPAGREHRRAWEQLITPRSMHVV
MAB_3449c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3974|M.avium_104 --------------------------------------------------
Mvan_5021|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
MSMEG_4472|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2728|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1146|M.marinum_M --------------------------------------------------
MUL_0911|M.ulcerans_Agy99 --------------------------------------------------
TH_3412|M.thermoresistible__bu --------------------------------------------------
Mb3755|M.bovis_AF2122/97 RREFVADDLRVLATRIAGRSVGLVLSGGAARACAHLGVLEELEAAGVTVD
Rv3728|M.tuberculosis_H37Rv RREFVADDLRVLATRIAGRSVGLVLSGGAARACAHLGVLEELEAAGVTVD
MAB_3449c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3974|M.avium_104 --------------------------------------------------
Mvan_5021|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
MSMEG_4472|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2728|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1146|M.marinum_M --------------------------------------------------
MUL_0911|M.ulcerans_Agy99 --------------------------------------------------
TH_3412|M.thermoresistible__bu --------------------------------------------------
Mb3755|M.bovis_AF2122/97 RFAGTSMGAIIAALAASGLDAAGVDAQIYEHFVRKSHGDYTLPSKGLIRG
Rv3728|M.tuberculosis_H37Rv RFAGTSMGAIIAALAASGLDAAGVDAQIYEHFVRKSHGDYTLPSKGLIRG
MAB_3449c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3974|M.avium_104 --------------------------------------------------
Mvan_5021|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
MSMEG_4472|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2728|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1146|M.marinum_M --------------------------------------------------
MUL_0911|M.ulcerans_Agy99 --------------------------------------------------
TH_3412|M.thermoresistible__bu --------------------------------------------------
Mb3755|M.bovis_AF2122/97 KRTQSTLRTIFGDHLVEELPKHFRCVSVDLLARRPVVHRQGPLADVVGCS
Rv3728|M.tuberculosis_H37Rv KRTQSTLRTIFGDHLVEELPKHFRCVSVDLLARRPVVHRQGPLADVVGCS
MAB_3449c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3974|M.avium_104 --------------------------------------------------
Mvan_5021|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
MSMEG_4472|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2728|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1146|M.marinum_M --------------------------------------------------
MUL_0911|M.ulcerans_Agy99 --------------------------------------------------
TH_3412|M.thermoresistible__bu --------------------------------------------------
Mb3755|M.bovis_AF2122/97 MRLPFLYAPLPYGGTLHVDGGVLDNVPVTTLVGKDGPLIAVNVASGGNPS
Rv3728|M.tuberculosis_H37Rv MRLPFLYAPLPYGGTLHVDGGVLDNVPVTTLVGKDGPLIAVNVASGGNPS
MAB_3449c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3974|M.avium_104 --------------------------------------------------
Mvan_5021|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
MSMEG_4472|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2728|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1146|M.marinum_M --------------------------------------------------
MUL_0911|M.ulcerans_Agy99 --------------------------------------------------
TH_3412|M.thermoresistible__bu --------------------------------------------------
Mb3755|M.bovis_AF2122/97 PASGGHRRGKPRVPGLTDTLLRTMTISSAMASEKVLAQADLVIKPNPIGV
Rv3728|M.tuberculosis_H37Rv PASGGHRRGKPRVPGLTDTLLRTMTISSAMASEKVLAQADLVIKPNPIGV
MAB_3449c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3974|M.avium_104 --------------------------------------------------
Mvan_5021|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
MSMEG_4472|M.smegmatis_MC2_155 --------------------------------
Mflv_2728|M.gilvum_PYR-GCK --------------------------------
MMAR_1146|M.marinum_M --------------------------------
MUL_0911|M.ulcerans_Agy99 --------------------------------
TH_3412|M.thermoresistible__bu --------------------------------
Mb3755|M.bovis_AF2122/97 GLMEYHQIDRAREAGRIAAREALPQIMELVHG
Rv3728|M.tuberculosis_H37Rv GLMEYHQIDRAREAGRIAAREALPQIMELVHG
MAB_3449c|M.abscessus_ATCC_199 --------------------------------
MAV_3974|M.avium_104 --------------------------------
Mvan_5021|M.vanbaalenii_PYR-1 --------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------