For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
STDIIYSDATGLAELIRSRQLSPVEVVQAHLDRIEAVNPKINAIVTVAERALAQARSAEAAVMRGDDLPP LHGVPFTVKDSIDTADVLTQRGSPIFAGRIPQTDAVSVARLKAAGAIVLAKTNLPEFSYSTETDNLLTGR SNNPWNLDRTPGGSSGGESAAIAAGLSPLGLGTDLAISVRGPAAQTGIAALKATHGRIPMTGVWPRVPRR FWHVGPMARTVRDLALAYELLAGPDGADAFAISPVRFDAGTAAVDSLRVGWLVEPGFGPIDAEVAATVRA AAEALSSIGLAVEPVRIPALERDFALDVFMRLHVMELKPAFAEATAGRGDEELFAISKRILATPDTSIQD FVEAEQAGERLRDGFADYFSRYDALLAPVLPVPAHEHRAKEFDINGQTVDATYIMGATVPFNVTGLPALS LRFGTSSAGLPINVQLISTWLAESTVLNLASLLESVSPVRNQHPGI*
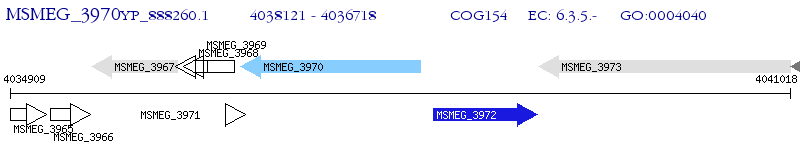
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3970 | - | - | 100% (467) | glutamyl-tRNA(Gln) amidotransferase subunit A |
| M. smegmatis MC2 155 | MSMEG_2986 | - | 5e-60 | 37.06% (456) | amidohydrolase, AtzE family protein |
| M. smegmatis MC2 155 | MSMEG_2497 | - | 5e-60 | 35.60% (455) | putative amidase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3200 | - | 2e-63 | 34.09% (484) | amidase |
| M. gilvum PYR-GCK | Mflv_4246 | gatA | 1e-49 | 30.30% (495) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. tuberculosis H37Rv | Rv3175 | - | 2e-63 | 34.09% (484) | amidase |
| M. leprae Br4923 | MLBr_01702 | gatA | 1e-49 | 31.55% (485) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. abscessus ATCC 19977 | MAB_3341c | gatA | 8e-52 | 31.03% (493) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. marinum M | MMAR_2765 | - | 1e-65 | 36.97% (476) | amidase |
| M. avium 104 | MAV_3859 | gatA | 1e-48 | 31.20% (500) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. thermoresistible (build 8) | TH_2639 | - | 2e-49 | 31.24% (493) | PUTATIVE glutamyl-tRNA (Gln) amidotransferase (subunit A), GatA |
| M. ulcerans Agy99 | MUL_1939 | gatA | 3e-49 | 31.49% (489) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. vanbaalenii PYR-1 | Mvan_2115 | gatA | 2e-46 | 31.03% (506) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_01702|M.leprae_Br4923 ---MIGSLHEIIRLDASTLAAKIVAKELSSVEITQACLDQIEATDDTYRA
MAV_3859|M.avium_104 -------MNEIIRSDAATLAARIAAKELSSVEATQACLDQIAATDERYHA
MUL_1939|M.ulcerans_Agy99 -------MTDLIRSDAATLAAKIAAKEVSATELTQACLDQIEATDDRYHA
Mflv_4246|M.gilvum_PYR-GCK -------MSELIKLDAATLADKISSKEVSSAEVTQAFLDQIAATDGDYHA
Mvan_2115|M.vanbaalenii_PYR-1 -------MTDLIKLDAATLAAKIAAREVSATEVTQACLDQIAATDAEYHA
TH_2639|M.thermoresistible__bu -MSQGKSTDDLIRLDAATLGAKIAAKEISSTELTQACLDRIAATDDRYNA
MAB_3341c|M.abscessus_ATCC_199 -------MTDLIRQDAAALAALIHSGEVSSVEVTRAHLDQIASTDETYRA
MSMEG_3970|M.smegmatis_MC2_155 ------MSTDIIYSDATGLAELIRSRQLSPVEVVQAHLDRIEAVNPKINA
MMAR_2765|M.marinum_M -------MTELCHLSAHELVGLMSTGSVSCREVIQQHLARIRAVNPALNA
Mb3200|M.bovis_AF2122/97 MAMSAKASDDIAWLPATAQLAVLAAKKVSSAELVELYLSRIDTYNASLNA
Rv3175|M.tuberculosis_H37Rv MAMSAKASDDIAWLPATAQLAVLAAKKVSSAELVELYLSRIDTYNASLNA
:: * : : .:* * . * :* : : .*
MLBr_01702|M.leprae_Br4923 FLHVAAEKALSAAAAVDK----AVAAGGQLSSTLAGVPLALKDVFTTVDM
MAV_3859|M.avium_104 FLHVAADEALGAAAVVDE----AVAAGERLPSPLAGVPLALKDVFTTVDM
MUL_1939|M.ulcerans_Agy99 FLHIGAHEALSAAAAVDT----ALAAGERLPSALAGVPLALKDVFTTVDM
Mflv_4246|M.gilvum_PYR-GCK FLHVGAEQALAAAQTVDR----AVAAGEHLPSPLAGVPLALKDVFTTTDM
Mvan_2115|M.vanbaalenii_PYR-1 FLHVAGDQALAAAATVDKAIHEATAAGERLPSPLAGVPLALKDVFTTTDM
TH_2639|M.thermoresistible__bu FLHVAANKALSAAARVDA----AVAAGEELPSPLAGVPLALKDVFTTIDM
MAB_3341c|M.abscessus_ATCC_199 FLHVAGEQALAAAKDVDD----AIAAGS-VPSGLAGVPLALKDVFTTRDM
MSMEG_3970|M.smegmatis_MC2_155 IVTVA--ERALAQARSAE----AAVMRGDDLPPLHGVPFTVKDSIDTADV
MMAR_2765|M.marinum_M LVEADDPERCLRQADHAD----ECVARGAPLGAAHGLPVVIKDVMQVAGL
Mb3200|M.bovis_AF2122/97 IVTVDP-DAARRVAKRSD----AARARGDELGPLHGLPITVKDSYETAGM
Rv3175|M.tuberculosis_H37Rv IVTVDP-DAARRVAKRSD----AARARGDELGPLHGLPITVKDSYETAGM
:: . *:*..:** . .:
MLBr_01702|M.leprae_Br4923 PTTCGSKILQGWHSPYDATVTTRLRAAGIPILGKTNMDEFAMGSSTENSA
MAV_3859|M.avium_104 PTTCGSKILQGWRSPYDATVTTKLRAAGIPILGKTNMDEFAMGSSTENSA
MUL_1939|M.ulcerans_Agy99 PTTCGSKILEGWRSPYDATLTLRLRAAGIPILGKTNMDEFAMGSSTENSA
Mflv_4246|M.gilvum_PYR-GCK PTTCGSKVLEGWTSPYDATVTAKLRSAGIPILGKTNMDEFAMGSSTENSA
Mvan_2115|M.vanbaalenii_PYR-1 PTTCGSKILEGWTSPYDATVTAKLRAAGIPILGKTNMDEFAMGSSTENSA
TH_2639|M.thermoresistible__bu PTTCGSKILDGWQAPYDATVTVALRAAGIPILGKTNLDEFAMGSSTENSA
MAB_3341c|M.abscessus_ATCC_199 PTTCGSKILENWVPPYDATVTARLRGAGIPILGKTNMDEFAMGSSTENSA
MSMEG_3970|M.smegmatis_MC2_155 LTQRGSPIFAGRIPQTDAVSVARLKAAGAIVLAKTNLPEFSYSTETDNLL
MMAR_2765|M.marinum_M ACSGGSPGLR-AVADTDATVVSRLRAQGAIVLGMTNVPEMSRGGESNNNL
Mb3200|M.bovis_AF2122/97 RTTCGRRDLADYVPTQDAEAVARLRRAGAIIMGKTNMPTGNQDVQASNPV
Rv3175|M.tuberculosis_H37Rv RTTCGRRDLADYVPTQDAEAVARLRRAGAIIMGKTNMPTGNQDVQASNPV
* : . ** . *: * ::. **: . .:.*
MLBr_01702|M.leprae_Br4923 YGPTRNPWNVDRVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTA
MAV_3859|M.avium_104 YGPTRNPWNVERVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTA
MUL_1939|M.ulcerans_Agy99 YGPTRNPWNLDRVPGGSGGGSAAALAAYQAPLAIGSDTGGSIRQPAALTA
Mflv_4246|M.gilvum_PYR-GCK YGPTRNPWDTERVPGGSGGGSAAALAAFQAPLAIGTDTGGSIRQPAALTA
Mvan_2115|M.vanbaalenii_PYR-1 YGPTRNPWNVDCVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTA
TH_2639|M.thermoresistible__bu FGPTRNPWDVNRVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTA
MAB_3341c|M.abscessus_ATCC_199 YGPTRNPWDVERVPGGSGGGSAAALAAFQAPLAIGTDTGGSIRQPAALTA
MSMEG_3970|M.smegmatis_MC2_155 TGRSNNPWNLDRTPGGSSGGESAAIAAGLSPLGLGTDLAISVRGPAAQTG
MMAR_2765|M.marinum_M YGRTNNPYDLTRTPGGSSGGSAALVAAGGAALSVGSDGGGSIRQPCHNTG
Mb3200|M.bovis_AF2122/97 FGRTNNPWDAARTSGGSAGGGAAATAAGLTSFDYGSEIGGSTRIPAHYCG
Rv3175|M.tuberculosis_H37Rv FGRTNNPWDAARTSGGSAGGGAAATAAGLTSFDYGSEIGGSTRIPAHYCG
* :.**:: ..***.** :* ** :.: *:: . * * *. .
MLBr_01702|M.leprae_Br4923 TVGVKPTYGTVSRYGLVACASS---------LDQGGPCARTVLDTALLHA
MAV_3859|M.avium_104 TVGVKPTYGTVSRYGLVACASS---------LDQGGPCARTVLDTALLHQ
MUL_1939|M.ulcerans_Agy99 TVGVKPTYGTVSRYGLVACASS---------LDQGGPCARTVLDTAMLHQ
Mflv_4246|M.gilvum_PYR-GCK TVGVKPTYGTVSRYGLVACASS---------LDQGGPCARTVLDTALLHQ
Mvan_2115|M.vanbaalenii_PYR-1 TVGVKPTYGTVSRYGLIACASS---------LDQGGPCARTVLDTALLHQ
TH_2639|M.thermoresistible__bu TVGVKPTYGTVSRYGLVACASS---------LDQGGPCGRTVLDTALLHQ
MAB_3341c|M.abscessus_ATCC_199 TVGVKPTYGTVSRFGLVACASS---------LDQGGPCARTVLDTALLHQ
MSMEG_3970|M.smegmatis_MC2_155 IAALKATHGRIPMTG-VWPRVP-------RRFWHVGPMARTVRDLALAYE
MMAR_2765|M.marinum_M IAGLKPTHGRIPRTGSVFGDALGV----FSPFVCYGPLARSVRDLFLGLS
Mb3200|M.bovis_AF2122/97 LYGHKSTWRSVPLVGHIPSAPGNPGRWGQADMACAGVQVRGARDIIPALE
Rv3175|M.tuberculosis_H37Rv LYGHKSTWRSVPLVGHIPSAPGNPGRWGQADMACAGVQVRGARDIIPALE
. *.* :. * : : * * . *
MLBr_01702|M.leprae_Br4923 VIAGHDARDSTSVEAAVPDIVGAAKAGESGDLHGVRVGVVRQLR-GEGYQ
MAV_3859|M.avium_104 VIAGHDIRDSTSVDAPVPDVVGAARAGAAGDLKGVRVGVVKQLR-GEGYQ
MUL_1939|M.ulcerans_Agy99 VIAGHDAKDSTSLETEIPDVVGAAKAGASGDLRGVRIGVVKQLR-GDGYQ
Mflv_4246|M.gilvum_PYR-GCK VIAGHDPLDSTSVEAPVPDVVAAARTGAGGDLTGVRIGVVKQLRSGEGYQ
Mvan_2115|M.vanbaalenii_PYR-1 VIAGHDPRDSTSVNAAVPDVVGAARAGARGDLKGVRIGVVKQLRSGEGYQ
TH_2639|M.thermoresistible__bu VIAGHDAKDSTSLTAPVPDVVAAARAGASGDLKGVRVGVVRQLR-GEGIQ
MAB_3341c|M.abscessus_ATCC_199 VIAGHDPRDSTSVDEPVPDVVAAARAGASGDLKGVRVGVVTQLR-GDGYQ
MSMEG_3970|M.smegmatis_MC2_155 LLAGPDGADAFAISPVRFDAG-------TAAVDSLRVGWLVEPG-FGPID
MMAR_2765|M.marinum_M IMNGPDLADPYTAPAPLGSPD-------DVELGTLRVAAYLDDG-ISPPS
Mb3200|M.bovis_AF2122/97 ATVGPMRADG-GFSYALAPPR-------AGALKDFRVAVWAEDP-HCPID
Rv3175|M.tuberculosis_H37Rv ATVGPMRADG-GFSYALAPPR-------AGALKDFRVAVWAEDP-HCPID
* * : .*:. : .
MLBr_01702|M.leprae_Br4923 SGVLASFQAAVEQLIALGATVSEVDCPHFDYALAAYYLILPSEVSSNLAR
MAV_3859|M.avium_104 PGVLASFEAAVEQLTALGAEVSEVDCPHFEYALAAYYLILPSEVSSNLAR
MUL_1939|M.ulcerans_Agy99 PGVLASFEAAVAQLTALGAEVSEVDCPHFDHALAAYYLILPSEVSSNLAR
Mflv_4246|M.gilvum_PYR-GCK AGVLASFNAAVDQLTALGAEVTEVDCPHFDYSLPAYYLILPSEVSSNLAK
Mvan_2115|M.vanbaalenii_PYR-1 PGVLASFTAAVEQLTALGAEVSEVDCPHFDHSLAAYYLILPSEVSSNLAK
TH_2639|M.thermoresistible__bu PGVLASFDAAVEQLAALGAEVIEVDCPNIDHALAAYYLIMPSEVSSNLAR
MAB_3341c|M.abscessus_ATCC_199 PGVMASFDAAVEQLTALGADVVEVSCPHFEYALPAYYLILPSEVSSNLAR
MSMEG_3970|M.smegmatis_MC2_155 AEVAATVRAAAEALSSI-GLAVEPVR------------IPALERDFALDV
MMAR_2765|M.marinum_M EEVTAVVTAAVEALREV-VAVIDHAV------------PPCLNRTTDLLW
Mb3200|M.bovis_AF2122/97 ADVRRAMDDAVAALRAAGAHVVEQPA------------TIPVDMAVSHNI
Rv3175|M.tuberculosis_H37Rv ADVRRAMDDAVAALRAAGAHVVEQPA------------TIPVDMAVSHNI
* . *. * . : :
MLBr_01702|M.leprae_Br4923 FDAMRYGLRVGDDGSRSAEEVIALTRAAGFGPEVKRRIMIGAYALSAGYY
MAV_3859|M.avium_104 FDAMRYGLRIGDDGSHSAEEVMALTRAAGFGPEVKRRIMIGTYALSAGYY
MUL_1939|M.ulcerans_Agy99 FDAMRYGLRIGDDGTHSAEEVMAMTRAAGFGPEVKRRIMIGAYALSAGYY
Mflv_4246|M.gilvum_PYR-GCK FDGMRYGLRAGDDGTHSAEEVMALTRAAGFGPEVKRRIMIGAYALSAGYY
Mvan_2115|M.vanbaalenii_PYR-1 FDGMRYGLRVGDDGTTSAEEVMAMTRAAGFGAEVKRRIMIGTYALSAGYY
TH_2639|M.thermoresistible__bu FDAMRYGLRVGDDGTRSAEEVMALTRAAGFGPEVKRRIMIGTYALSAGYY
MAB_3341c|M.abscessus_ATCC_199 FDAMRYGMRVGDDGTHSAEEVMALTRAAGFGPEVKRRIMIGTYALSAGYY
MSMEG_3970|M.smegmatis_MC2_155 FMRLHV----MELKPAFAEATAGRGD---EELFAISKRILATPDTSIQDF
MMAR_2765|M.marinum_M TSIFLG----GDRGRGFEEDLGAIGTTAPSEELAEFTAQARQVDFSLTEA
Mb3200|M.bovis_AF2122/97 FQSLVFGAFAVDRSTLSPASAAALGLRAVRHPRGEAANALGATLQSHRAW
Rv3175|M.tuberculosis_H37Rv FQSLVFGAFAVDRSTLSPASAAALGLRAVRHPRGEAANALGATLQSHRAW
: : . *
MLBr_01702|M.leprae_Br4923 DSYYSQAQKVRTLIARDLDKAYRSVDVLVSPATPTTAFSLG---------
MAV_3859|M.avium_104 DAYYNQAQKVRTLIARDLDAAYESVDVVVSPATPTTAFGLG---------
MUL_1939|M.ulcerans_Agy99 DAYYNQAQKIRTLIARDLDEAYQSVDVLVSPATPTTAFPLG---------
Mflv_4246|M.gilvum_PYR-GCK DAYYNQAQKVRTLIARDLDAAYQKVDVLVSPATPTTAFRLG---------
Mvan_2115|M.vanbaalenii_PYR-1 DAYYNQAQKVRTLIARDLDEAYRSVDVLVSPATPSTAFRLG---------
TH_2639|M.thermoresistible__bu DAYYNQAQKVRTLVARDFDAAYEKVDVLVSPTTPTTAFPLG---------
MAB_3341c|M.abscessus_ATCC_199 DAYYGRAQKVRTLIKRDLERAYEQVDVLVTPATPTTAFRLG---------
MSMEG_3970|M.smegmatis_MC2_155 VEAEQAGERLR----DGFADYFSRYDALLAPVLPVPAHEHR-------AK
MMAR_2765|M.marinum_M RRRLVDIDHYR----IQMLAFMADYDVIIGPAMPTPAKPHH-------HG
Mb3200|M.bovis_AF2122/97 LFADAARHEMR----DRWAGFFNEFDVLLLPVTPTPAPLHHNKDHDRLGR
Rv3175|M.tuberculosis_H37Rv LFADAARHEMR----DRWAGFFNEFDVLLLPVTPTPAPLHHNKDHDRLGR
.. * *.:: *. * .*
MLBr_01702|M.leprae_Br4923 --EKADDPLAMYLFDLCTLPLNLAGHCGMSVPSGLSSDDDLPVGLQIMAP
MAV_3859|M.avium_104 --EKVDDPLAMYLFDLCTLPLNLAGHCGMSVPSGLSPDDDLPVGLQIMAP
MUL_1939|M.ulcerans_Agy99 --EKVDDPLAMYLFDLCTLPLNLAGHCGMSVPSGLSPDDGLPVGLQIMAP
Mflv_4246|M.gilvum_PYR-GCK --EKVDDPLSMYLFDLCTLPLNLAGHCGMSVPSGLSADDNLPVGLQIMAP
Mvan_2115|M.vanbaalenii_PYR-1 --EKVDDPLAMYLFDLCTLPLNLAGHCGMSVPSGLSADDNLPVGLQIMAP
TH_2639|M.thermoresistible__bu --EKVDHPLAMYLFDLCTLPLNLAGHCGMSVPSGLSPDDNLPVGLQIMAP
MAB_3341c|M.abscessus_ATCC_199 --EKVDDPLAMYQFDLCTLPLNLAGHCGMSVPSGLSADDGLPVGLQIMAP
MSMEG_3970|M.smegmatis_MC2_155 EFDINGQTVDATYIMGATVPFNVTGLPALSLRFGTSSA-GLPINVQLIST
MMAR_2765|M.marinum_M LVEIS----DFSHLM----VHNLTGWPAAVVRCGTSPG-GLPIGVQIAAR
Mb3200|M.bovis_AF2122/97 TIDVDGVSRSYWDQLKWNALANIAGTPATTMPITTTAT-GLPIGIQAMGP
Rv3175|M.tuberculosis_H37Rv TIDVDGVSRSYWDQLKWNALANIAGTPATTMPITTTAT-GLPIGIQAMGP
: *::* . : :. .**:.:* .
MLBr_01702|M.leprae_Br4923 ALADDRLYRVGAAYEAARGPLPSAI-----
MAV_3859|M.avium_104 ALADDRLYRVGAAYEAARGPLRSAI-----
MUL_1939|M.ulcerans_Agy99 ALADDRLYRVGAAYEAARGPLPSAI-----
Mflv_4246|M.gilvum_PYR-GCK ALADDRLYRVGAAYEAARGPLPTAL-----
Mvan_2115|M.vanbaalenii_PYR-1 ALADDRLYRVGAAYEAARGPLPTAL-----
TH_2639|M.thermoresistible__bu ALADDRLYRVGAAYEAARGPLPAAV-----
MAB_3341c|M.abscessus_ATCC_199 ALADDRLYRVGAAYETARGALPAAL-----
MSMEG_3970|M.smegmatis_MC2_155 WLAESTVLNLASLLESVSPVRNQHPGI---
MMAR_2765|M.marinum_M PWHDGTALAVASHLEAIFGG-WQAPGLLVT
Mb3200|M.bovis_AF2122/97 AGGDRTTVEFAALLTEVLGG-FRVPPL---
Rv3175|M.tuberculosis_H37Rv AGGDRTTVEFAALLTEVLGG-FRVPPL---
: ..: