For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTDLIKLDAATLAAKIAAREVSATEVTQACLDQIAATDAEYHAFLHVAGDQALAAAATVDKAIHEATAAG ERLPSPLAGVPLALKDVFTTTDMPTTCGSKILEGWTSPYDATVTAKLRAAGIPILGKTNMDEFAMGSSTE NSAYGPTRNPWNVDCVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTATVGVKPTYGTVSRYGLI ACASSLDQGGPCARTVLDTALLHQVIAGHDPRDSTSVNAAVPDVVGAARAGARGDLKGVRIGVVKQLRSG EGYQPGVLASFTAAVEQLTALGAEVSEVDCPHFDHSLAAYYLILPSEVSSNLAKFDGMRYGLRVGDDGTT SAEEVMAMTRAAGFGAEVKRRIMIGTYALSAGYYDAYYNQAQKVRTLIARDLDEAYRSVDVLVSPATPST AFRLGEKVDDPLAMYLFDLCTLPLNLAGHCGMSVPSGLSADDNLPVGLQIMAPALADDRLYRVGAAYEAA RGPLPTAL
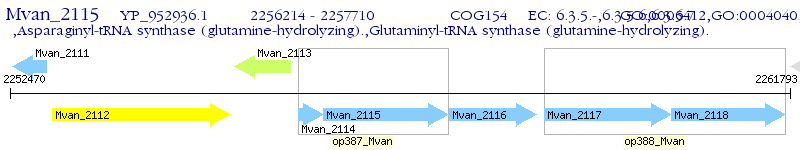
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2115 | gatA | - | 100% (498) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. vanbaalenii PYR-1 | Mvan_3824 | - | 1e-42 | 31.28% (486) | amidase |
| M. vanbaalenii PYR-1 | Mvan_5822 | - | 2e-34 | 29.65% (516) | amidase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3036c | gatA | 0.0 | 89.56% (498) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. gilvum PYR-GCK | Mflv_4246 | gatA | 0.0 | 89.36% (498) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. tuberculosis H37Rv | Rv3011c | gatA | 0.0 | 88.96% (498) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. leprae Br4923 | MLBr_01702 | gatA | 0.0 | 85.89% (496) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. abscessus ATCC 19977 | MAB_3341c | gatA | 0.0 | 84.34% (498) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. marinum M | MMAR_1702 | gatA | 0.0 | 88.15% (498) | glutamyl-tRNA (Gln) amidotransferase (subunit A), GatA |
| M. avium 104 | MAV_3859 | gatA | 0.0 | 88.55% (498) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. smegmatis MC2 155 | MSMEG_2365 | gatA | 0.0 | 88.93% (497) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. thermoresistible (build 8) | TH_2639 | - | 0.0 | 85.89% (496) | PUTATIVE glutamyl-tRNA (Gln) amidotransferase (subunit A), GatA |
| M. ulcerans Agy99 | MUL_1939 | gatA | 0.0 | 87.95% (498) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2115|M.vanbaalenii_PYR-1 ------MTDLIKLDAATLAAKIAAREVSATEVTQACLDQIAATDAEYHAF
Mflv_4246|M.gilvum_PYR-GCK ------MSELIKLDAATLADKISSKEVSSAEVTQAFLDQIAATDGDYHAF
MLBr_01702|M.leprae_Br4923 --MIGSLHEIIRLDASTLAAKIVAKELSSVEITQACLDQIEATDDTYRAF
MAV_3859|M.avium_104 ------MNEIIRSDAATLAARIAAKELSSVEATQACLDQIAATDERYHAF
Mb3036c|M.bovis_AF2122/97 ------MTDIIRSDAATLAAKIAIKEVSSTEITRACLDQIEATDETYHAF
Rv3011c|M.tuberculosis_H37Rv ------MTDIIRSDAATLAAKIAIKEVSSAEITRACLDQIEATDETYHAF
MMAR_1702|M.marinum_M ------MTDLIRSDAATLAAKIAAKEVSATELTQACLDQIEATDDRYHAF
MUL_1939|M.ulcerans_Agy99 ------MTDLIRSDAATLAAKIAAKEVSATELTQACLDQIEATDDRYHAF
MSMEG_2365|M.smegmatis_MC2_155 MS-----TELIRSDAATLGAKIAAKEVSSTEVTQAHLDQIAATDDRFHAF
TH_2639|M.thermoresistible__bu MSQGKSTDDLIRLDAATLGAKIAAKEISSTELTQACLDRIAATDDRYNAF
MAB_3341c|M.abscessus_ATCC_199 ------MTDLIRQDAAALAALIHSGEVSSVEVTRAHLDQIASTDETYRAF
::*: **::*. * *:*:.* *:* **:* :** :.**
Mvan_2115|M.vanbaalenii_PYR-1 LHVAGDQALAAAATVDKAIHEATAAGERLPSPLAGVPLALKDVFTTTDMP
Mflv_4246|M.gilvum_PYR-GCK LHVGAEQALAAAQTVDR----AVAAGEHLPSPLAGVPLALKDVFTTTDMP
MLBr_01702|M.leprae_Br4923 LHVAAEKALSAAAAVDK----AVAAGGQLSSTLAGVPLALKDVFTTVDMP
MAV_3859|M.avium_104 LHVAADEALGAAAVVDE----AVAAGERLPSPLAGVPLALKDVFTTVDMP
Mb3036c|M.bovis_AF2122/97 LHVAADEALAAAAAVDK----QVAAGEPLPSALAGVPLALKDVFTTSDMP
Rv3011c|M.tuberculosis_H37Rv LHVAADEALAAAAAIDK----QVAAGEPLPSALAGVPLALKDVFTTSDMP
MMAR_1702|M.marinum_M LHIGAHEALSAAAAVDT----ALAAGERLPSALAGVPLALKDVFTTVDMP
MUL_1939|M.ulcerans_Agy99 LHIGAHEALSAAAAVDT----ALAAGERLPSALAGVPLALKDVFTTVDMP
MSMEG_2365|M.smegmatis_MC2_155 LHVAADSALAEAARVDA----AVAAGEQLPSPLAGVPLALKDVFTTTDMP
TH_2639|M.thermoresistible__bu LHVAANKALSAAARVDA----AVAAGEELPSPLAGVPLALKDVFTTIDMP
MAB_3341c|M.abscessus_ATCC_199 LHVAGEQALAAAKDVDD----AIAAGS-VPSGLAGVPLALKDVFTTRDMP
**:....**. * :* *** :.* ************** ***
Mvan_2115|M.vanbaalenii_PYR-1 TTCGSKILEGWTSPYDATVTAKLRAAGIPILGKTNMDEFAMGSSTENSAY
Mflv_4246|M.gilvum_PYR-GCK TTCGSKVLEGWTSPYDATVTAKLRSAGIPILGKTNMDEFAMGSSTENSAY
MLBr_01702|M.leprae_Br4923 TTCGSKILQGWHSPYDATVTTRLRAAGIPILGKTNMDEFAMGSSTENSAY
MAV_3859|M.avium_104 TTCGSKILQGWRSPYDATVTTKLRAAGIPILGKTNMDEFAMGSSTENSAY
Mb3036c|M.bovis_AF2122/97 TTCGSKILEGWRSPYDATLTARLRAAGIPILGKTNMDEFAMGSSTENSAY
Rv3011c|M.tuberculosis_H37Rv TTCGSKILEGWRSPYDATLTARLRAAGIPILGKTNMDEFAMGSSTENSAY
MMAR_1702|M.marinum_M TTCGSKILEGWRSPYDATLTLRLRAAGIPILGKTNMDEFAMGSSTENSAY
MUL_1939|M.ulcerans_Agy99 TTCGSKILEGWRSPYDATLTLRLRAAGIPILGKTNMDEFAMGSSTENSAY
MSMEG_2365|M.smegmatis_MC2_155 TTCGSKILEGWQSPYDATVTVKLRAAGIPILGKTNMDEFAMGSSTENSAY
TH_2639|M.thermoresistible__bu TTCGSKILDGWQAPYDATVTVALRAAGIPILGKTNLDEFAMGSSTENSAF
MAB_3341c|M.abscessus_ATCC_199 TTCGSKILENWVPPYDATVTARLRGAGIPILGKTNMDEFAMGSSTENSAY
******:*:.* .*****:* **.**********:*************:
Mvan_2115|M.vanbaalenii_PYR-1 GPTRNPWNVDCVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTAT
Mflv_4246|M.gilvum_PYR-GCK GPTRNPWDTERVPGGSGGGSAAALAAFQAPLAIGTDTGGSIRQPAALTAT
MLBr_01702|M.leprae_Br4923 GPTRNPWNVDRVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTAT
MAV_3859|M.avium_104 GPTRNPWNVERVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTAT
Mb3036c|M.bovis_AF2122/97 GPTRNPWNLDRVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTAT
Rv3011c|M.tuberculosis_H37Rv GPTRNPWNLDRVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTAT
MMAR_1702|M.marinum_M GPTRNPWNLDRVPGGSGGGSAAALAAYQAPLAIGSDTGGSIRQPAALTAT
MUL_1939|M.ulcerans_Agy99 GPTRNPWNLDRVPGGSGGGSAAALAAYQAPLAIGSDTGGSIRQPAALTAT
MSMEG_2365|M.smegmatis_MC2_155 GPTRNPWNTERVPGGSGGGSAAALAAYQAPLAIGSDTGGSIRQPAALTAT
TH_2639|M.thermoresistible__bu GPTRNPWDVNRVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTAT
MAB_3341c|M.abscessus_ATCC_199 GPTRNPWDVERVPGGSGGGSAAALAAFQAPLAIGTDTGGSIRQPAALTAT
*******: : ***************:*******:***************
Mvan_2115|M.vanbaalenii_PYR-1 VGVKPTYGTVSRYGLIACASSLDQGGPCARTVLDTALLHQVIAGHDPRDS
Mflv_4246|M.gilvum_PYR-GCK VGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTALLHQVIAGHDPLDS
MLBr_01702|M.leprae_Br4923 VGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTALLHAVIAGHDARDS
MAV_3859|M.avium_104 VGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTALLHQVIAGHDIRDS
Mb3036c|M.bovis_AF2122/97 VGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTALLHQVIAGHDPRDS
Rv3011c|M.tuberculosis_H37Rv VGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTALLHQVIAGHDPRDS
MMAR_1702|M.marinum_M VGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTAMLHQVIAGHDAKDS
MUL_1939|M.ulcerans_Agy99 VGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTAMLHQVIAGHDAKDS
MSMEG_2365|M.smegmatis_MC2_155 VGVKPTYGTVSRYGLIACASSLDQGGPCARTVLDTALLHAVIAGHDARDS
TH_2639|M.thermoresistible__bu VGVKPTYGTVSRYGLVACASSLDQGGPCGRTVLDTALLHQVIAGHDAKDS
MAB_3341c|M.abscessus_ATCC_199 VGVKPTYGTVSRFGLVACASSLDQGGPCARTVLDTALLHQVIAGHDPRDS
************:**:************.*******:** ****** **
Mvan_2115|M.vanbaalenii_PYR-1 TSVNAAVPDVVGAARAGARGDLKGVRIGVVKQLRSGEGYQPGVLASFTAA
Mflv_4246|M.gilvum_PYR-GCK TSVEAPVPDVVAAARTGAGGDLTGVRIGVVKQLRSGEGYQAGVLASFNAA
MLBr_01702|M.leprae_Br4923 TSVEAAVPDIVGAAKAGESGDLHGVRVGVVRQLRG-EGYQSGVLASFQAA
MAV_3859|M.avium_104 TSVDAPVPDVVGAARAGAAGDLKGVRVGVVKQLRG-EGYQPGVLASFEAA
Mb3036c|M.bovis_AF2122/97 TSVDAEVPDVVGAARAGAVGDLRGVRVGVVRQLHGGEGYQPGVLASFEAA
Rv3011c|M.tuberculosis_H37Rv TSVDAEVPDVVGAARAGAVGDLRGVRVGVVRQLHGGEGYQPGVLASFEAA
MMAR_1702|M.marinum_M TSLETEIPDVVGAAKAGASGDLRGVRIGVVKQLRG-DGYQPGVLASFEAA
MUL_1939|M.ulcerans_Agy99 TSLETEIPDVVGAAKAGASGDLRGVRIGVVKQLRG-DGYQPGVLASFEAA
MSMEG_2365|M.smegmatis_MC2_155 TSVKADVPDVVAAARAGATGDLKGVRVGVVKQLRG-DGYQPGVLQSFNAA
TH_2639|M.thermoresistible__bu TSLTAPVPDVVAAARAGASGDLKGVRVGVVRQLRG-EGIQPGVLASFDAA
MAB_3341c|M.abscessus_ATCC_199 TSVDEPVPDVVAAARAGASGDLKGVRVGVVTQLRG-DGYQPGVMASFDAA
**: :**:*.**::* *** ***:*** **:. :* *.**: ** **
Mvan_2115|M.vanbaalenii_PYR-1 VEQLTALGAEVSEVDCPHFDHSLAAYYLILPSEVSSNLAKFDGMRYGLRV
Mflv_4246|M.gilvum_PYR-GCK VDQLTALGAEVTEVDCPHFDYSLPAYYLILPSEVSSNLAKFDGMRYGLRA
MLBr_01702|M.leprae_Br4923 VEQLIALGATVSEVDCPHFDYALAAYYLILPSEVSSNLARFDAMRYGLRV
MAV_3859|M.avium_104 VEQLTALGAEVSEVDCPHFEYALAAYYLILPSEVSSNLARFDAMRYGLRI
Mb3036c|M.bovis_AF2122/97 VEQLTALGAEVSEVDCPHFDHALAAYYLILPSEVSSNLARFDAMRYGLRV
Rv3011c|M.tuberculosis_H37Rv VEQLTALGAEVSEVDCPHFDHALAAYYLILPSEVSSNLARFDAMRYGLRV
MMAR_1702|M.marinum_M VAQLTALGAEVSEVDCPHFDHALAAYYLILPSEVSSNLARFDAMRYGLRI
MUL_1939|M.ulcerans_Agy99 VAQLTALGAEVSEVDCPHFDHALAAYYLILPSEVSSNLARFDAMRYGLRI
MSMEG_2365|M.smegmatis_MC2_155 VEQLTALGAEVTEVDCPHFDHAMAAYYLILPSEVSSNLARFDAMRFGLRV
TH_2639|M.thermoresistible__bu VEQLAALGAEVIEVDCPNIDHALAAYYLIMPSEVSSNLARFDAMRYGLRV
MAB_3341c|M.abscessus_ATCC_199 VEQLTALGADVVEVSCPHFEYALPAYYLILPSEVSSNLARFDAMRYGMRV
* ** **** * **.**::::::.*****:*********:**.**:*:*
Mvan_2115|M.vanbaalenii_PYR-1 GDDGTTSAEEVMAMTRAAGFGAEVKRRIMIGTYALSAGYYDAYYNQAQKV
Mflv_4246|M.gilvum_PYR-GCK GDDGTHSAEEVMALTRAAGFGPEVKRRIMIGAYALSAGYYDAYYNQAQKV
MLBr_01702|M.leprae_Br4923 GDDGSRSAEEVIALTRAAGFGPEVKRRIMIGAYALSAGYYDSYYSQAQKV
MAV_3859|M.avium_104 GDDGSHSAEEVMALTRAAGFGPEVKRRIMIGTYALSAGYYDAYYNQAQKV
Mb3036c|M.bovis_AF2122/97 GDDGTRSAEEVMAMTRAAGFGPEVKRRIMIGTYALSAGYYDAYYNQAQKV
Rv3011c|M.tuberculosis_H37Rv GDDGTRSAEEVMAMTRAAGFGPEVKRRIMIGTYALSAGYYDAYYNQAQKV
MMAR_1702|M.marinum_M GDDGTHSAEEVMAMTRAAGFGPEVKRRIMIGAYALSAGYYDAYYNQAQKV
MUL_1939|M.ulcerans_Agy99 GDDGTHSAEEVMAMTRAAGFGPEVKRRIMIGAYALSAGYYDAYYNQAQKI
MSMEG_2365|M.smegmatis_MC2_155 GDDGTHSAEEVMALTRAAGFGPEVKRRIMIGTYALSAGYYDAYYNQAQKV
TH_2639|M.thermoresistible__bu GDDGTRSAEEVMALTRAAGFGPEVKRRIMIGTYALSAGYYDAYYNQAQKV
MAB_3341c|M.abscessus_ATCC_199 GDDGTHSAEEVMALTRAAGFGPEVKRRIMIGTYALSAGYYDAYYGRAQKV
****: *****:*:*******.*********:*********:**.:***:
Mvan_2115|M.vanbaalenii_PYR-1 RTLIARDLDEAYRSVDVLVSPATPSTAFRLGEKVDDPLAMYLFDLCTLPL
Mflv_4246|M.gilvum_PYR-GCK RTLIARDLDAAYQKVDVLVSPATPTTAFRLGEKVDDPLSMYLFDLCTLPL
MLBr_01702|M.leprae_Br4923 RTLIARDLDKAYRSVDVLVSPATPTTAFSLGEKADDPLAMYLFDLCTLPL
MAV_3859|M.avium_104 RTLIARDLDAAYESVDVVVSPATPTTAFGLGEKVDDPLAMYLFDLCTLPL
Mb3036c|M.bovis_AF2122/97 RTLIARDLDAAYRSVDVLVSPTTPTTAFRLGEKVDDPLAMYLFDLCTLPL
Rv3011c|M.tuberculosis_H37Rv RTLIARDLDAAYRSVDVLVSPTTPTTAFRMGEKVDDPLAMYLFDLCTLPL
MMAR_1702|M.marinum_M RTLIARDLDEAYQSVDVLVSPATPTTAFPLGEKVDDPLAMYLFDLCTLPL
MUL_1939|M.ulcerans_Agy99 RTLIARDLDEAYQSVDVLVSPATPTTAFPLGEKVDDPLAMYLFDLCTLPL
MSMEG_2365|M.smegmatis_MC2_155 RTLIARDLDRAYENVDVLVSPATPTTAFGLGEKVDDPLAMYLFDLCTLPL
TH_2639|M.thermoresistible__bu RTLVARDFDAAYEKVDVLVSPTTPTTAFPLGEKVDHPLAMYLFDLCTLPL
MAB_3341c|M.abscessus_ATCC_199 RTLIKRDLERAYEQVDVLVTPATPTTAFRLGEKVDDPLAMYQFDLCTLPL
***: **:: **..***:*:*:**:*** :***.*.**:** ********
Mvan_2115|M.vanbaalenii_PYR-1 NLAGHCGMSVPSGLSADDNLPVGLQIMAPALADDRLYRVGAAYEAARGPL
Mflv_4246|M.gilvum_PYR-GCK NLAGHCGMSVPSGLSADDNLPVGLQIMAPALADDRLYRVGAAYEAARGPL
MLBr_01702|M.leprae_Br4923 NLAGHCGMSVPSGLSSDDDLPVGLQIMAPALADDRLYRVGAAYEAARGPL
MAV_3859|M.avium_104 NLAGHCGMSVPSGLSPDDDLPVGLQIMAPALADDRLYRVGAAYEAARGPL
Mb3036c|M.bovis_AF2122/97 NLAGHCGMSVPSGLSPDDGLPVGLQIMAPALADDRLYRVGAAYEAARGPL
Rv3011c|M.tuberculosis_H37Rv NLAGHCGMSVPSGLSPDDGLPVGLQIMAPALADDRLYRVGAAYEAARGPL
MMAR_1702|M.marinum_M NLAGHCGMSVPSGLSPDDGLPVGLQIMAPALADDRLYRVGAAYEAARGPL
MUL_1939|M.ulcerans_Agy99 NLAGHCGMSVPSGLSPDDGLPVGLQIMAPALADDRLYRVGAAYEAARGPL
MSMEG_2365|M.smegmatis_MC2_155 NLAGHCGMSVPSGLSPDDGLPVGLQIMAPALADDRLYRVGAAYEAARGPL
TH_2639|M.thermoresistible__bu NLAGHCGMSVPSGLSPDDNLPVGLQIMAPALADDRLYRVGAAYEAARGPL
MAB_3341c|M.abscessus_ATCC_199 NLAGHCGMSVPSGLSADDGLPVGLQIMAPALADDRLYRVGAAYETARGAL
***************.**.*************************:***.*
Mvan_2115|M.vanbaalenii_PYR-1 PTAL
Mflv_4246|M.gilvum_PYR-GCK PTAL
MLBr_01702|M.leprae_Br4923 PSAI
MAV_3859|M.avium_104 RSAI
Mb3036c|M.bovis_AF2122/97 LSAI
Rv3011c|M.tuberculosis_H37Rv LSAI
MMAR_1702|M.marinum_M PSAI
MUL_1939|M.ulcerans_Agy99 PSAI
MSMEG_2365|M.smegmatis_MC2_155 PSAL
TH_2639|M.thermoresistible__bu PAAV
MAB_3341c|M.abscessus_ATCC_199 PAAL
:*: