For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
QPTMNAVIERPDDLTGEWLTAALGAGRVTDFSFERIGTGQMSECYRVTLRYADGTDGPASVVLKVAATDP NSRQTGLALGLYEREVRFYTEIAPQLGAGPIAPCYHSAFDPQTGAFDLLLGDAAPATVGNELQGATVEQA TLALVQLGHVHGAAHASSRGAAIADWLVREAPVSQPMLAGLYAAFVERYADMITPQQREVCERLVESFDS YLAAEDERPKGLVHGDYRLDNMLFGDPGADRPLTAVDWQTVTWGPALTDVAYFVGSSLPSEVRREHYDAL LRAYHEALGPDAPLTLDQVREDVRRQSFFGVVMAIVSSMLVERTERGDQMFMTMLDRNCSHVLDTGALDI LAPPVPVAPLQPAAEDDDAHTPGDEPLWNESWYFDFVDTAAGFGGWIRLGLIPNQDHSWINVLVCGPDLP TVALNDFHAALPTDVARVCGDGVELSLQRGEPLQTYRVTASGQGQAYDDPAALLRGEAGTPVDVTLDLTW TTVGTPYQYRITPRYEIPCTVTGTVTVGETVHPLEAVVGQRDHSWGVRDWWSMEWVWSALHLDDGTHLHG VDLRIPNMPPLGVGYTQRDGELVELQSVTAQHTLGDDDLPVTTTLTLQPGDIEATVDIQGHAPVKLVSAE GKVSQFPRAWATVRTADGRTGVGWLEWNRNLG*
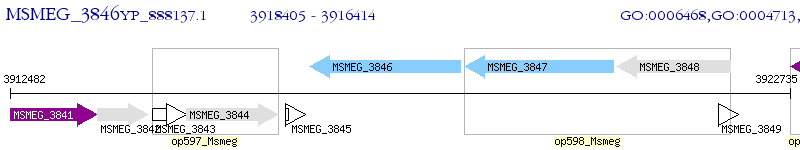
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3846 | - | - | 100% (663) | phosphotransferase enzyme family protein |
| M. smegmatis MC2 155 | MSMEG_3876 | - | 3e-36 | 31.62% (351) | phosphotransferase enzyme family protein, putative |
| M. smegmatis MC2 155 | MSMEG_4885 | - | 3e-13 | 27.57% (341) | hypothetical protein MSMEG_4885 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3155 | - | 0.0 | 64.65% (662) | hypothetical protein Mflv_3155 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4589c | - | 4e-06 | 43.75% (48) | putative phosphotransferase |
| M. marinum M | MMAR_3019 | - | 0.0 | 63.01% (657) | hypothetical protein MMAR_3019 |
| M. avium 104 | MAV_2467 | - | 0.0 | 65.35% (658) | phosphotransferase enzyme family protein |
| M. thermoresistible (build 8) | TH_2511 | - | 0.0 | 66.02% (668) | PUTATIVE protein of unknown function DUF227 |
| M. ulcerans Agy99 | MUL_1045 | - | 1e-14 | 29.18% (257) | hypothetical protein MUL_1045 |
| M. vanbaalenii PYR-1 | Mvan_3374 | - | 0.0 | 65.91% (657) | aminoglycoside phosphotransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3155|M.gilvum_PYR-GCK MADRPLLYLGRVCSLTRPQ---LIERPADLTAEWLTDIL-----GHGQVE
Mvan_3374|M.vanbaalenii_PYR-1 ---------------MRPR---LIERPPDLTAEWLTAVI-----GQGTVT
MSMEG_3846|M.smegmatis_MC2_155 ------------MQPTMNA---VIERPDDLTGEWLTAAL-----GAGRVT
TH_2511|M.thermoresistible__bu ------------VQPTRNR---VIERPTDLTTHWLTEKLTAGTPGAGRVT
MMAR_3019|M.marinum_M ------------MNDEGDQNAGIIEEPGDLTASWLTEKI-----GAGDVT
MAV_2467|M.avium_104 ------------MHAETEQ---VIERPSDLSASWLAAVI-----GTGPIA
MUL_1045|M.ulcerans_Agy99 ----MIALARLATHLGRGLGRVATDAVLRNARAFPRRIHDLDPPALSEIM
MAB_4589c|M.abscessus_ATCC_199 -------MHKDVSSDSDGPTGDAASLDADAIPTSLSEYLN-----RELGE
.
Mflv_3155|M.gilvum_PYR-GCK SFTVDRIGTGQMSECYRVGIRYAGGG-VGPASVVLKVAAAEPSSRQTGLA
Mvan_3374|M.vanbaalenii_PYR-1 EFTVDRIGTGQMSECYRVALSYADDG-AGPATVVLKVAAADPSSRQTGLA
MSMEG_3846|M.smegmatis_MC2_155 DFSFERIGTGQMSECYRVTLRYA-DGTDGPASVVLKVAATDPNSRQTGLA
TH_2511|M.thermoresistible__bu DFTVERIGTGQMSECYRVTLSWTGDGADRPASVVLKVAASDPTSRQTGVS
MMAR_3019|M.marinum_M EFAVERIGTGQMSECYRVQLTYAVP--GGPSLVVLKVAATDPMSRQTGQA
MAV_2467|M.avium_104 DFSVERIGTGQMSECYRIRLRYAQADARGPESVVLKVAATDPVSRQTGLA
MUL_1045|M.ulcerans_Agy99 GRRVSSVSVLDDSSGTTSRARLALSGNDVPESVFVKMAAATAATRFIGEL
MAB_4589c|M.abscessus_ATCC_199 QVSVRRLSAGHSNLTYVVTGSSGTQWILRRPPFGRLQAGAHDVMREYRVL
. :.. . . . *.: *
Mflv_3155|M.gilvum_PYR-GCK MGLYEREVRFYTDIAPSLG-GPVAPCHHTAYDPDTGAFDLVLADAAPASV
Mvan_3374|M.vanbaalenii_PYR-1 MGLYEREVRFYTDIAPGLG-GPVAPCHHGAYDPHTGAFDLLLADAAPALV
MSMEG_3846|M.smegmatis_MC2_155 LGLYEREVRFYTEIAPQLGAGPIAPCYHSAFDPQTGAFDLLLGDAAPATV
TH_2511|M.thermoresistible__bu LGLYEREVRFYADIAPRLTGGPIAPCYHAAYDPDSGAFDLLLGDAAPAQV
MMAR_3019|M.marinum_M LGLYEREVRFYAEVAPRLG-GPIAPCYHAAVDAQTGAFDLLLGDAGPALV
MAV_2467|M.avium_104 LGLYEREVRFYGDVAPRLG-GPIAPCYHAAVDTSTGVFDLLLGDAGPAVV
MUL_1045|M.ulcerans_Agy99 GRLAETEVSFYRDLSPELA-PDAPTAYGCAFDPLTGRFVLVLEDLAAPAE
MAB_4589c|M.abscessus_ATCC_199 DALSTAQVRVPRVTLASTD----AEIFGAPFYLMERQDGEVIRDVSPEWF
* :* . . . . . :: * ..
Mflv_3155|M.gilvum_PYR-GCK G-----DEIRGATAEQASLAMTQLGLVHGPLLGSEA-LEGADWLNRESPV
Mvan_3374|M.vanbaalenii_PYR-1 G-----DEIRGATVEEAMLAVTQIGLVHGPLLNDEA-LAGADWLNRESPV
MSMEG_3846|M.smegmatis_MC2_155 G-----NELQGATVEQATLALVQLGHVHGAAHASSRGAAIADWLVREAPV
TH_2511|M.thermoresistible__bu G-----DEIRGATPEQAKLALAELGKVHAPLLGDEQ-LGGAEWLNRDAPI
MMAR_3019|M.marinum_M G-----DEIAGATVAQAQLAVSQLGRLHGPLLGDAA-LAEAPWLNRDSPL
MAV_2467|M.avium_104 G-----DEIAGATIEQARLGVVELGRLHGPLLGDIS-LAQAPWLNRDAPL
MUL_1045|M.ulcerans_Agy99 GTCDFSDTLHPFDNDQAALVVELLARLHATYWVRLN-HGGVRPFGWRYAA
MAB_4589c|M.abscessus_ATCC_199 VG-----SARRELVLDLAVALAEIHNADPTRLVEAKLGRESGYLARQLKT
: : : : . . :
Mflv_3155|M.gilvum_PYR-GCK NQGLVAALYAGYIDRYSDQIAPEHRVVCERLVASFDAYMAAEDAAG-GPR
Mvan_3374|M.vanbaalenii_PYR-1 NQGLISALYAGYIDRYREQIAPEHRNVCERFVESFDAYMAAENDAD-TPR
MSMEG_3846|M.smegmatis_MC2_155 SQPMLAGLYAAFVERYADMITPQQREVCERLVESFDSYLAAEDE---RPK
TH_2511|M.thermoresistible__bu DQALLAQLYAGFVERYGNLVSDAHRLVCERLIAAFDDYLAAEAAG--RPH
MMAR_3019|M.marinum_M NQAMVNGLYAGFLDRYGEQIDPRSRMVCERLVAAFDGYLAQESSRG-AIQ
MAV_2467|M.avium_104 NQAMIAPLYAGFVERYGDQIAPEHRAVCERLVAAFDGYLAQEP----GVR
MUL_1045|M.ulcerans_Agy99 SRDPSVPLVPTMLRASWRRLGGHAAALLPGRDFVFEHYRAITGLIDAPPH
MAB_4589c|M.abscessus_ATCC_199 WGG----QWESIKELPTGRDLQDHTTITAWLTENYPGD---------RPA
: :
Mflv_3155|M.gilvum_PYR-GCK GLVHGDFRLDNMLFGQDGSDRPLTVVDWQTVTWGPAFTDVSYFLGGSLPI
Mvan_3374|M.vanbaalenii_PYR-1 GLVHGDYRLDNMLFGQDGAHRPLTVVDWQTVTWGPAFSDVAYFLGCALPV
MSMEG_3846|M.smegmatis_MC2_155 GLVHGDYRLDNMLFGDPGADRPLTAVDWQTVTWGPALTDVAYFVGSSLPS
TH_2511|M.thermoresistible__bu GLVHGDFRLDNMLFGDPHAARPLTAVDWQTVAWGPALTDVAYFLGCALKT
MMAR_3019|M.marinum_M GLVHGDYRLDNLLFGAAGAERPLTVVDWQTVSWGPALTDLAYFLGCALQP
MAV_2467|M.avium_104 GLVHGDYRLDNMLFGTAGAERALTVVDWQTVSWGPALTDLAYFLGCALPT
MUL_1045|M.ulcerans_Agy99 TVLHGDAHPGNWYFRSSGGPLAAGLLDWQAIRLGHPARDLAYTLITGMTT
MAB_4589c|M.abscessus_ATCC_199 AVVHGDYKLDNVLVDP-ATARITAVLDWEMATVGDPLADLGYLLYMTPEP
::*** : .* . :**: * . *:.* :
Mflv_3155|M.gilvum_PYR-GCK EERRAHYDELLAAYHRALGEATGVSLDDVREGVRHQSFFGVLMAIVSSML
Mvan_3374|M.vanbaalenii_PYR-1 EQRREHYDALLAAYHGALGPTAGVSIDEVREGVRRQSFFGVLMSIVSPML
MSMEG_3846|M.smegmatis_MC2_155 EVRREHYDALLRAYHEALGPDAPLTLDQVREDVRRQSFFGVVMAIVSSML
TH_2511|M.thermoresistible__bu EDRRAHYDELLRAYHDALGPEPALTLDEVRDRVRRQTFTGVVMAIVPAML
MMAR_3019|M.marinum_M NERRAHYDDLLRTYHDALGPQAPITLEQVADEVRRQSFFGVMMAIVSSML
MAV_2467|M.avium_104 EDRREHYDALLRAYHEALGPQPPITLADVADGVRRQSFFGVMMAIVSSML
MUL_1045|M.ulcerans_Agy99 TDRRVTERELLDVYRRALAAAGGPELDREEPWLRYR--------------
MAB_4589c|M.abscessus_ATCC_199 GGE-VAMSELTGSVTAQEGCPSHTEIAQAYREARSG--------------
. * . : *
Mflv_3155|M.gilvum_PYR-GCK VGRTERGDEMFMALIARHCQHVLDTDALAILPAPSTPEPLQPNADDEGRH
Mvan_3374|M.vanbaalenii_PYR-1 VERTERGDEMFMAMIARHCQHVIDTGALAVLPEPGTPEPLQPNADDEGRH
MSMEG_3846|M.smegmatis_MC2_155 VERTERGDQMFMTMLDRNCSHVLDTGALDILAPPVPVAPLQPAAEDDDAH
TH_2511|M.thermoresistible__bu VERTERGDEMFLTLLGRHCDHVLDTGALDILPPPAAPEPLRPEPADEYPH
MMAR_3019|M.marinum_M VERTDRGDRLFMTMLHRHCEHVLDTDALATLPDAAPPEPLCPGEQDEMAH
MAV_2467|M.avium_104 VERTDRGDQMFMTMLRRHCDHVLDTDALATLPAASAPEPLQPSPEDELAH
MUL_1045|M.ulcerans_Agy99 --------------QAAACAYVATT-------------------------
MAB_4589c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_3155|M.gilvum_PYR-GCK APTEEPLWSESWYFDFAAPSQDIGGWIRLGLMPNEGHAWVNGLLCGPGMP
Mvan_3374|M.vanbaalenii_PYR-1 SPTDEALWSESWYFDFADPGQQLGGWIRLGLIPNQDHAWINGLWCAPGGP
MSMEG_3846|M.smegmatis_MC2_155 TPGDEPLWNESWYFDFVDTAAGFGGWIRLGLIPNQDHSWINVLVCGPDLP
TH_2511|M.thermoresistible__bu RPTDEPLWNESWYFDFVDAEQGVGGWVRLGLMPNEGVAWVNALLCGPDMP
MMAR_3019|M.marinum_M EPTDEPLWSESWYADFVDAAQGLGGWFRLGRIANQQKAWVHALLCGPDMA
MAV_2467|M.avium_104 DPTTEPLWSESWYADFADPAQGLGGWFRLGLVANQQTAWVHALVCGPDMP
MUL_1045|M.ulcerans_Agy99 ------------------IAAGLG-----------GMQAADIALTGLDLT
MAB_4589c|M.abscessus_ATCC_199 ---DQSYWTP-------------------------EDLVYYQVLGGWKLA
. .
Mflv_3155|M.gilvum_PYR-GCK TIAALDFEAPLPEEHTRVHGQDIELEQTVIEPLQTYQVTVRGRGQAHDDP
Mvan_3374|M.vanbaalenii_PYR-1 TIAVLDFEAPLPEDHTHVAADGIELDLEVLEPLSSYRVVLRGRGEAHDDP
MSMEG_3846|M.smegmatis_MC2_155 TVALNDFHAALPTDVARVCGDGVELSLQRGEPLQTYRVTASGQGQAYDDP
TH_2511|M.thermoresistible__bu TIALTEWEAPLPADPFRTRTGTAELDMEATDPLRSYRVTVRGRGQSYDDP
MMAR_3019|M.marinum_M TVAVADIEVALPDDPWAVRTGDIELRHHATAPLQTYAVQLRARGQAYEDP
MAV_2467|M.avium_104 TVAV-DAQVPLPPDPWTVRTDTFEITHSAGAPLRSYRVDVRALAQAYADP
MUL_1045|M.ulcerans_Agy99 VAALHDLQT----------------------------------------V
MAB_4589c|M.abscessus_ATCC_199 TLLEVSYQRHLAG-------------------------------------
. . .
Mflv_3155|M.gilvum_PYR-GCK AALLRGEPGRDVEVTMDLTWTSVGQPYQYRVSPRYEIPCTVTGTVSADGH
Mvan_3374|M.vanbaalenii_PYR-1 AALLRGEAGRPVDVSMDLTWSTVGRPYQYRMSPRYEIPCTVTGTVVADGR
MSMEG_3846|M.smegmatis_MC2_155 AALLRGEAGTPVDVTLDLTWTTVGTPYQYRITPRYEIPCTVTGTVTVGET
TH_2511|M.thermoresistible__bu AVLLAGGAGEPVELTFDLEWTSAGTPYQYRITPRFEIPCTVSGTVTAGDR
MMAR_3019|M.marinum_M SALLRGEPGDPVEVTMNLVWTTNGVPYRYRLTSRYEIPCTVAGTVTVDGT
MAV_2467|M.avium_104 AALLRGEPGTPVAMTMNLDWHTDGTPYKYAMTTRYEIPCTVTGSVTIGDT
MUL_1045|M.ulcerans_Agy99 AALRAADLG--------------GTSWAWRPG------MARSARIRPGAS
MAB_4589c|M.abscessus_ATCC_199 ---------------------TTDDPFFAELDEGVPRLLSVARSLIPAAG
. .: : : :
Mflv_3155|M.gilvum_PYR-GCK TYEFDAVVGQRDHSWAARDWWAMDWVWSAFHLEDGTHIHGVEILIPGAPP
Mvan_3374|M.vanbaalenii_PYR-1 TYAIAGVPGQRDHSWAARDWWSMDWMWSALHLDDGTHIHGVEILIPGMPP
MSMEG_3846|M.smegmatis_MC2_155 VHPLEAVVGQRDHSWGVRDWWSMEWVWSALHLDDGTHLHGVDLRIPNMPP
TH_2511|M.thermoresistible__bu SYTVTAVPGQRDHSWGVRDWWGMEWVWSALHLDDGTRLHGVDVRIPDMGP
MMAR_3019|M.marinum_M HYRVDAVPGQRDHSWGVRDWWSMDWMWSALHLDDGTHLHGVNIQLPGTPA
MAV_2467|M.avium_104 AYRIESVAGQRDHSWGVRDWWSMDWIWSALHLDDGTHLHGVNIRIPGAPA
MUL_1045|M.ulcerans_Agy99 ARRCPMVWSTSSHILGVSRHFAMGAW------------------------
MAB_4589c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_3155|M.gilvum_PYR-GCK MSIGYLQRAGEPLVELSSVSAQTTFADNDLPQSAVLDYG--DLRVDVTVR
Mvan_3374|M.vanbaalenii_PYR-1 LAVGYVQRDGESLVELAKVSARTTFADNGLPISAQVDYAPGDITATIDVR
MSMEG_3846|M.smegmatis_MC2_155 LGVGYTQR-DGELVELQSVTAQHTLGDDDLPVTTTLTLQPGDIEATVDIQ
TH_2511|M.thermoresistible__bu IGIGYIQPPDGPLLELSGVRARETVAGTGLPVATTIELTPGDVSVDIDIQ
MMAR_3019|M.marinum_M FSVGYVQDSGGNLTELQAVNFREAFGSNGLPQNTTLILKPGDITANIDVR
MAV_2467|M.avium_104 FSIGYTQGADGEVTELQTVDSRESFGDNGLPLDATLELNPGEITAQVDVR
MUL_1045|M.ulcerans_Agy99 --------------------------------------------------
MAB_4589c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_3155|M.gilvum_PYR-GCK GHAPVLLTSADGRLSQFPRAWATVTTDDGRTGAGWIEWNRNLAQPDG
Mvan_3374|M.vanbaalenii_PYR-1 GHAPVLLTSPDGRHSRFPRAWGEVTTADGRTGVGWIEWNRSQD----
MSMEG_3846|M.smegmatis_MC2_155 GHAPVKLVSAEGKVSQFPRAWATVRTADGRTGVGWLEWNRNLG----
TH_2511|M.thermoresistible__bu GHAPVRLTAPDGRVSHFPRAWCRVTTADGRTGVGWVEWNRVQT----
MMAR_3019|M.marinum_M AHAPVRLNASDWRVSHFPRAWVAVVTTDGRSGVGWMEWNRNQV----
MAV_2467|M.avium_104 GQAPVRLVAADGRVSQFPRVWAAVSTGDGRRGVGWLEWNRNLGERIG
MUL_1045|M.ulcerans_Agy99 -----------------------------------------------
MAB_4589c|M.abscessus_ATCC_199 -----------------------------------------------