For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KAISVEQFGGPDVLELSEVPDPRPAPGEVLISIEASGVGYVDVMAREGHYHAFTEAGFVPGLEIAGVVSE LGDGVDAGWLGRRVFAMPMTGGGYAEYVAVSSDQLIPLPDGVSARSAVALGMNALVAKIAVERMGSAPGE SVYVRGAGGGIGIMAVQICNARGAHVTATTSSALRGERLRALGAEHVVDRHASGESDSRSYDLIVDTVAG PQLGSYVAKLKPNGRYILCGGVGGAPAEDFGRVLLENFHRSPTFSAFSLNSVENPAIVTTGTELFAGADR GAITPVVHCELPLADAASAHAELEAGRVFGKVVLIP*
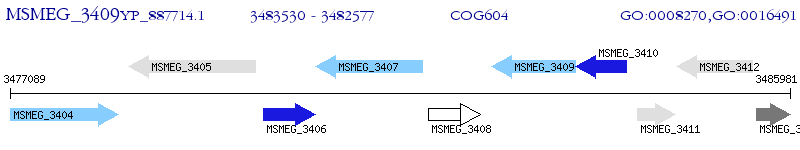
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3409 | - | - | 100% (317) | oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_4032 | - | 9e-39 | 36.17% (329) | zinc-binding alcohol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_4064 | - | 3e-38 | 35.87% (329) | zinc-binding alcohol dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1947c | fadB5 | 1e-35 | 30.95% (336) | oxidoreductase FADB5 |
| M. gilvum PYR-GCK | Mflv_1189 | - | 2e-38 | 33.64% (321) | alcohol dehydrogenase |
| M. tuberculosis H37Rv | Rv3777 | - | 2e-35 | 32.06% (315) | oxidoreductase |
| M. leprae Br4923 | MLBr_00118 | - | 6e-33 | 31.60% (307) | putative oxidireductase |
| M. abscessus ATCC 19977 | MAB_1112c | - | 2e-41 | 35.06% (328) | putative quinone oxidoreductase |
| M. marinum M | MMAR_2814 | fadB5 | 7e-39 | 32.74% (336) | oxidoreductase FadB5 |
| M. avium 104 | MAV_3325 | - | 4e-35 | 32.52% (329) | quinone oxidoreductase |
| M. thermoresistible (build 8) | TH_1665 | - | 5e-37 | 33.33% (333) | PROBABLE OXIDOREDUCTASE |
| M. ulcerans Agy99 | MUL_2935 | fadB5 | 2e-37 | 33.04% (339) | oxidoreductase FadB5 |
| M. vanbaalenii PYR-1 | Mvan_5615 | - | 8e-37 | 33.33% (312) | alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1189|M.gilvum_PYR-GCK -------MAE---TPDQLNWQSVPDPTP-GPGEVLIRVHAAGVNRADLLQ
Mvan_5615|M.vanbaalenii_PYR-1 --MHAIVAQP---SSEGLTWQSVPDIAP-APGEVLIRVAAAGVNRADLLQ
TH_1665|M.thermoresistible__bu ---MRAISVQ---TSGQLSWVEVPDIEP-GESEVLIAVHTAGINRADLLQ
Rv3777|M.tuberculosis_H37Rv MTIMRAVVAE---SSDRLVWQEVPDVSA-GPGEVLIKVAASGVNRADVLQ
MLBr_00118|M.leprae_Br4923 ---MHAIVAE---SANQMVWREVPDVTA-GPGEVLIEVAASGVNRADVLQ
MAB_1112c|M.abscessus_ATCC_199 ---MYAITLNGFGDPEVMQWTQVADLPAPGPGEVTIDVVAAGVNRADVMQ
MSMEG_3409|M.smegmatis_MC2_155 ---MKAISVEQFGGPDVLELSEVPDPRP-APGEVLISIEASGVGYVDVMA
MAV_3325|M.avium_104 ---MHAIEVSETGGPEVLRYVETPTPTA-GPGQVLIKAEAIGVNYIDTYF
MMAR_2814|M.marinum_M ---MRAVVITRHGDPSVLQVQQRPDPPPPGPGQLRISVRAAGVNFADHLA
MUL_2935|M.ulcerans_Agy99 ---MRAVVITKHGDPSVLQVQQRPDPPPPGPGQLRIAVRAAGVNFADHLA
Mb1947c|M.bovis_AF2122/97 ---MRAVVITKHGDPSVLQVRQRPDPPPPGPGQLRVAVRAAGVNFADHLA
. : . . . . .:: : : *:. *
Mflv_1189|M.gilvum_PYR-GCK AAGKYPPPPGASEIIGLEASGTIAAVGDGVTEWSEGQRVCALLAGGG-YA
Mvan_5615|M.vanbaalenii_PYR-1 AAGKYPPPPGASDILGLEVSGTIAAIGEGVTKWSQGQPVCALLAGGG-YA
TH_1665|M.thermoresistible__bu AAGKYPPPPGASEILGLEVSGTVAAVGAGVSGWTVGQPVCALLAGGG-YA
Rv3777|M.tuberculosis_H37Rv AAGKYPPPPGVSDIIGLEVSGIVAAVGPGVTEWSAGQEVCALLAGGG-YA
MLBr_00118|M.leprae_Br4923 VAGKYPHPPGASAIIGMEVAGVVAAVSSGVTEWSVGQEVCALLAGGG-YA
MAB_1112c|M.abscessus_ATCC_199 RLGLYPPPPGVSEIPGLEVSGHIAEVGPAVQGWVPGDAVCALMAGGG-YA
MSMEG_3409|M.smegmatis_MC2_155 REGHYHAFTEAGFVPGLEIAGVVSELGDGVDAGWLGRRVFAMPMTGGGYA
MAV_3325|M.avium_104 RSGQYP--RELPFIIGQELCGTVAAAGDGVDGFKVGDRVVSAAATGG-YA
MMAR_2814|M.marinum_M RVGLYPDAPKLPAVVGYEVAGTVEAVGAGVDESRIGERVLAGTRFGG-YS
MUL_2935|M.ulcerans_Agy99 RVGLYPDAPKLPAVVGYEVAGTVEAAGAGVDESRIGERVLAGTRFGG-YS
Mb1947c|M.bovis_AF2122/97 RVGLYPDAPKLPAVVGYEVAGTVEAVGDGVDPNRVGERVLAGTRFGG-YC
* * : * * .* : . .* * * : ** *.
Mflv_1189|M.gilvum_PYR-GCK EYVAVPAGQVLPVPDGVPLDHAAGLPEVACTVWSNLVMTARLRAPELLLI
Mvan_5615|M.vanbaalenii_PYR-1 EFVAVPAGQVLPIPDGVPVPHAAGLPEVACTVWSNVVMAAGLTAPELLLV
TH_1665|M.thermoresistible__bu EYVAVPAEQVMPIPESVEIRYAAALPEVACTVWSNLVMTAGLSAGEWVLI
Rv3777|M.tuberculosis_H37Rv EYVAVPADQVLPIPPSVNLVDSAALPEVACTVWSNLVMTAHLRPGQLVLI
MLBr_00118|M.leprae_Br4923 EYVAVPASQVMPIPKGVNVVDSAGIPEVACTVWSNLVMAAHLSAGQLLLL
MAB_1112c|M.abscessus_ATCC_199 ERVNVAATQVLPVPRGVSITAAAALPETAATVWSNVVMTGGLRSGQTLLI
MSMEG_3409|M.smegmatis_MC2_155 EYVAVSSDQLIPLPDGVSARSAVALGMNALVAKIAVERMG-SAPGESVYV
MAV_3325|M.avium_104 EYATAPASLTAHVPDGVAADVAASALLKGLTAHYLLTSVYPVQAGDTVLV
MMAR_2814|M.marinum_M EIVNVTATDAVVLPEGLSFEQGAAVPVNYATAWAALHGYGSLRAGERVLV
MUL_2935|M.ulcerans_Agy99 EIVNVTATDSVVLPEGLSFEQGAAVPVNYATAWAALHGYGSLRAGERVLV
Mb1947c|M.bovis_AF2122/97 EIVNVAATDSVVLPDALSFEQGAAVPVNYATAWAALHGYGSLRAGERVLI
* . ..: :* .: ... .. : . : : :
Mflv_1189|M.gilvum_PYR-GCK HGGASGIGTHAIQVARALNCRVAVTAGSRNKLDLCAELGAEITIDYHHE-
Mvan_5615|M.vanbaalenii_PYR-1 HGGASGIGTHAIQVAKALNCRVAVTAGSRNKLDLCAELGADITIDYHNE-
TH_1665|M.thermoresistible__bu HGGASGVGSHAVQVARALDARVAVTAGTRHKLELCRELGAEVTICYREE-
Rv3777|M.tuberculosis_H37Rv HGGASGIGSHAIQVVRALAARVAITAGSPEKLELCRDLGAQITINYRDE-
MLBr_00118|M.leprae_Br4923 HGGTSGIGSHGIQVARALGAKVAVTAGSEEKLEFCRALGAQITINYRDE-
MAB_1112c|M.abscessus_ATCC_199 HGGGSGIGTHAIQVGRALGANVAVTAGTQFKLDRCRELGAHTLINYREQ-
MSMEG_3409|M.smegmatis_MC2_155 RGAGGGIGIMAVQICNARGAHVTATTSSALRGERLRALGAEHVVDR----
MAV_3325|M.avium_104 HAGAGGVGLILTQWAHLLGARVITTVSTPEKARRSAQAGANEVLSYPDDA
MMAR_2814|M.marinum_M HAAAGGVGIAAVQFAKAAGAQVHGTAS-PGKHQRLAELGVDRAIDYR---
MUL_2935|M.ulcerans_Agy99 HAAAGGVGIAAVQFAKAAGAQVHGTAS-PGKHQRLAELGVDRAIDYR---
Mb1947c|M.bovis_AF2122/97 HAAAGGVGIAAVQFAKAAKAEVHGTAS-PQKHQKLAEFGVDRAIDYR---
:.. .*:* * . ..* *.. : *.. :
Mflv_1189|M.gilvum_PYR-GCK -DFVEVLRGADSDHPGADVILDIMGASYLDRNIDALATGGRLVIIGMQGG
Mvan_5615|M.vanbaalenii_PYR-1 -DYVGVVRDAG----GADVILDIMGAAYLDRNIETLADGGRLVIIGMQGG
TH_1665|M.thermoresistible__bu -DFVERVRAETDG-AGADVVLDIMGAAYLGRNVDALATDGRLVVIGMQGG
Rv3777|M.tuberculosis_H37Rv -DFVARLKQETDG-SGADIILDIMGASYLDRNIDALATDGQLIVIGMQGG
MLBr_00118|M.leprae_Br4923 -DFVARLQQQN---GGADVILDIMGAAYLDRNIDALAIDGQLIVIGLQGG
MAB_1112c|M.abscessus_ATCC_199 -DFPAVVNAAY---SGANVILDIVGGGYLAKNVEALAENGHLTIIGLQGG
MSMEG_3409|M.smegmatis_MC2_155 -----HASGESDS-RSYDLIVDTVAGPQLGSYVAKLKPNGRYILCGGVGG
MAV_3325|M.avium_104 DEFGNRIRALTDG-AGVAAVYDGVGASTFDASLASLAVRGTLALFGAASG
MMAR_2814|M.marinum_M -----RDGWWKGL-DSYDLVLDALGGTSLRRSYELLRPGGRLVGYGVSNM
MUL_2935|M.ulcerans_Agy99 -----RDGWWKGL-DSYDLVLDALGDTYLRRSYELLRPGGRLVGYGVSNM
Mb1947c|M.bovis_AF2122/97 -----RDGWWQGL-GPYDVVLDALGGTSLRRSYTLLRPGGRLVGYGISNM
: * :. : * * * .
Mflv_1189|M.gilvum_PYR-GCK TKAEINLGKLLAKRGSVMATALRSRPVTGRGSKS------------VIVS
Mvan_5615|M.vanbaalenii_PYR-1 AKAELNIGKLLTKRGSVIATALRSRPVHGRGGKA------------DIVA
TH_1665|M.thermoresistible__bu RRAELDLGKLVAKRAAVFGTALRGRPVTGPHGKA------------DIVR
Rv3777|M.tuberculosis_H37Rv VKAELNLGKLLTKRARVIGTTLRARPVSGPHGKA------------AIAQ
MLBr_00118|M.leprae_Br4923 VKGELNLGKVLSKRARIIGSTLRARPVNGPNSKS------------EIVA
MAB_1112c|M.abscessus_ATCC_199 AAAELNLGALLFKRGSVHATNLRRRPEQGPGSKA------------EIIA
MSMEG_3409|M.smegmatis_MC2_155 APAEDFGRVLLENFHRSPTFSAFSLNSVEN----------------PAIV
MAV_3325|M.avium_104 PVPPFDPQRLNAAGSVFLTRPSLAHFIRTG----------------QEFS
MMAR_2814|M.marinum_M QEGEKRSLRRVAPHALSMLRGFNLMKQLEESKTVIGLNMLRLWDDRGTLE
MUL_2935|M.ulcerans_Agy99 QEGEKRSLRRVAPHALSMLRGFNLMKQLEESKTVIGLNMLRLWDDRGTLE
Mb1947c|M.bovis_AF2122/97 QHGEKRSMRRVAPHALSMLRGFNLMKQLEESKTVIGLNMLRLWDDRRTLE
Mflv_1189|M.gilvum_PYR-GCK EVIENVWPLVADGQVRPVIGAEFPISEAQAAHELLASGDVSGKVILRVDV
Mvan_5615|M.vanbaalenii_PYR-1 RVGAELWPLVADGQVRPVIGAEFPIAEAQAAHELLASGDVSGKVLLRVDV
TH_1665|M.thermoresistible__bu AVVENVWPMVASDQVRPVVGAEFPIQEAQAAHELLASGDVYGKVLLCVED
Rv3777|M.tuberculosis_H37Rv AVAASVWPMIAANRVRPVIGTRLPIQQAAQAHELMLSGKTFGKILLTV--
MLBr_00118|M.leprae_Br4923 AVTASIWPMIADGSVRPIIGARMAVQQAADAHQLLLSGKVSGKIVLTVAG
MAB_1112c|M.abscessus_ATCC_199 ELRQHLWPLIAEGTVAPVVSAEVPVTDAAEAHRLLDSPETVGKVLLTVRR
MSMEG_3409|M.smegmatis_MC2_155 TTGTELFAGADRGAITPVVHCELPLADAASAHAELEAGRVFGKVVLIP--
MAV_3325|M.avium_104 WRAEELFAAIAAGDITVEVGGRYPLADAARAHRDLQGRKTTGSIVLLP--
MMAR_2814|M.marinum_M PWIAPLSEALDQGTAAPIVHAAIPFGEAPEAHRILAARENIGKVVLVP--
MUL_2935|M.ulcerans_Agy99 PWIAPLSEALGQGTAAPIVHAAIPFAEAPEAHRILAARENIGKVVLVP--
Mb1947c|M.bovis_AF2122/97 PWIAPLTKALNDGTILPIVHAIVPFAEAPEAHRILAARENVGKVVLVP--
: . : .. :* ** : . *.::*
Mflv_1189|M.gilvum_PYR-GCK -----------
Mvan_5615|M.vanbaalenii_PYR-1 -----------
TH_1665|M.thermoresistible__bu -----------
Rv3777|M.tuberculosis_H37Rv -----------
MLBr_00118|M.leprae_Br4923 SGTETLLSRHH
MAB_1112c|M.abscessus_ATCC_199 Q----------
MSMEG_3409|M.smegmatis_MC2_155 -----------
MAV_3325|M.avium_104 -----------
MMAR_2814|M.marinum_M -----------
MUL_2935|M.ulcerans_Agy99 -----------
Mb1947c|M.bovis_AF2122/97 -----------