For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
YAITLNGFGDPEVMQWTQVADLPAPGPGEVTIDVVAAGVNRADVMQRLGLYPPPPGVSEIPGLEVSGHIA EVGPAVQGWVPGDAVCALMAGGGYAERVNVAATQVLPVPRGVSITAAAALPETAATVWSNVVMTGGLRSG QTLLIHGGGSGIGTHAIQVGRALGANVAVTAGTQFKLDRCRELGAHTLINYREQDFPAVVNAAYSGANVI LDIVGGGYLAKNVEALAENGHLTIIGLQGGAAAELNLGALLFKRGSVHATNLRRRPEQGPGSKAEIIAEL RQHLWPLIAEGTVAPVVSAEVPVTDAAEAHRLLDSPETVGKVLLTVRRQ*
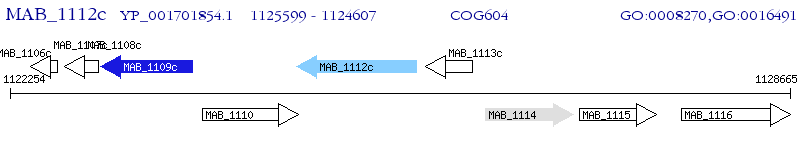
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1112c | - | - | 100% (330) | putative quinone oxidoreductase |
| M. abscessus ATCC 19977 | MAB_0207c | - | e-105 | 58.23% (328) | quinone oxidoreductase |
| M. abscessus ATCC 19977 | MAB_4603c | - | 5e-34 | 30.67% (326) | oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3806 | - | 4e-97 | 57.83% (313) | putative oxidoreductase |
| M. gilvum PYR-GCK | Mflv_1189 | - | 1e-100 | 59.25% (319) | alcohol dehydrogenase |
| M. tuberculosis H37Rv | Rv3777 | - | 8e-97 | 57.83% (313) | oxidoreductase |
| M. leprae Br4923 | MLBr_00118 | - | 1e-95 | 54.95% (313) | putative oxidireductase |
| M. marinum M | MMAR_5332 | - | 8e-96 | 59.11% (313) | oxidoreductase |
| M. avium 104 | MAV_0242 | - | 1e-101 | 58.47% (313) | quinone oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_6362 | - | 1e-100 | 58.47% (313) | quinone oxidoreductase |
| M. thermoresistible (build 8) | TH_1665 | - | 1e-96 | 55.32% (329) | PROBABLE OXIDOREDUCTASE |
| M. ulcerans Agy99 | MUL_0096 | - | 1e-94 | 58.47% (313) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_5615 | - | 3e-99 | 59.37% (315) | alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6362|M.smegmatis_MC2_155 ---MHAIVAS---VNGGLNWENVASIAP-ANGEVLIKVHTAGINRADLLQ
TH_1665|M.thermoresistible__bu ---MRAISVQ---TSGQLSWVEVPDIEP-GESEVLIAVHTAGINRADLLQ
Mb3806|M.bovis_AF2122/97 MTIMRAVVAE---SSDRLVWQEVPDVSA-GPGEVLIKVAASGVNRADVLQ
Rv3777|M.tuberculosis_H37Rv MTIMRAVVAE---SSDRLVWQEVPDVSA-GPGEVLIKVAASGVNRADVLQ
MMAR_5332|M.marinum_M MAFVHAIVAE---SADHLVWQQVPDVTA-GAGEVVIKVAAAGVNRADVLQ
MUL_0096|M.ulcerans_Agy99 MAFVHAIVAE---SADHLVWQQVPDVTA-GAGEVVIKVAAAGVNRADVLQ
MLBr_00118|M.leprae_Br4923 ---MHAIVAE---SANQMVWREVPDVTA-GPGEVLIEVAASGVNRADVLQ
MAV_0242|M.avium_104 MATMRAIVAE---SADQLSWQEVADVSA-GPGEVVVKVAAAGVNRADVLQ
Mflv_1189|M.gilvum_PYR-GCK -------MAE---TPDQLNWQSVPDPTP-GPGEVLIRVHAAGVNRADLLQ
Mvan_5615|M.vanbaalenii_PYR-1 --MHAIVAQP---SSEGLTWQSVPDIAP-APGEVLIRVAAAGVNRADLLQ
MAB_1112c|M.abscessus_ATCC_199 ---MYAITLNGFGDPEVMQWTQVADLPAPGPGEVTIDVVAAGVNRADVMQ
: * .*.. . . .** : * ::*:****::*
MSMEG_6362|M.smegmatis_MC2_155 AAGKYPPPPGASEVIGLEVSGTVEALGDGVSGWSVGQPVCALLAGGGYAE
TH_1665|M.thermoresistible__bu AAGKYPPPPGASEILGLEVSGTVAAVGAGVSGWTVGQPVCALLAGGGYAE
Mb3806|M.bovis_AF2122/97 AAGKYPPPPGVSDIIGLEVSGIVAAVGPGVTEWSAGQEVCALLAGGGYAE
Rv3777|M.tuberculosis_H37Rv AAGKYPPPPGVSDIIGLEVSGIVAAVGPGVTEWSAGQEVCALLAGGGYAE
MMAR_5332|M.marinum_M AAGKYPPPPGASEIIGMEVSGTITDIGDGVTEWSAGQEVCALLAGGGYAE
MUL_0096|M.ulcerans_Agy99 AAGKYPPPPGASEIIGMEVSGTITDIGDGVTEWSAGQEVCALLAGGGYAE
MLBr_00118|M.leprae_Br4923 VAGKYPHPPGASAIIGMEVAGVVAAVSSGVTEWSVGQEVCALLAGGGYAE
MAV_0242|M.avium_104 AAGKYPPPPGASETIGMEVSGVIAEVGDGVTEWSVGQEVCALLAGGGYAE
Mflv_1189|M.gilvum_PYR-GCK AAGKYPPPPGASEIIGLEASGTIAAVGDGVTEWSEGQRVCALLAGGGYAE
Mvan_5615|M.vanbaalenii_PYR-1 AAGKYPPPPGASDILGLEVSGTIAAIGEGVTKWSQGQPVCALLAGGGYAE
MAB_1112c|M.abscessus_ATCC_199 RLGLYPPPPGVSEIPGLEVSGHIAEVGPAVQGWVPGDAVCALMAGGGYAE
* ** ***.* *:*.:* : :. .* * *: ****:*******
MSMEG_6362|M.smegmatis_MC2_155 YVAVPAPQVLPVPDGVSLRDAAGLPEVACTVWSNVVMTAGLTAGHLLLVH
TH_1665|M.thermoresistible__bu YVAVPAEQVMPIPESVEIRYAAALPEVACTVWSNLVMTAGLSAGEWVLIH
Mb3806|M.bovis_AF2122/97 YVAVPADQVLPIPPSVNLVDSAALPEVACTVWSNLVMTAHLRPGQLVLIH
Rv3777|M.tuberculosis_H37Rv YVAVPADQVLPIPPSVNLVDSAALPEVACTVWSNLVMTAHLRPGQLVLIH
MMAR_5332|M.marinum_M YVAVPAGQVLPMPPGVSLIDAAALPEVACTVWSNLVMTAHLGPGQLVLIH
MUL_0096|M.ulcerans_Agy99 YVAVPAGQVLPMPPGVSLIDAAALPEVACTVWSNLVMTAHLGPGQLVLIH
MLBr_00118|M.leprae_Br4923 YVAVPASQVMPIPKGVNVVDSAGIPEVACTVWSNLVMAAHLSAGQLLLLH
MAV_0242|M.avium_104 YVAVPAGQLLPIPAGVDLVDAAGLPEVACTVWSNLVLIAGLRNGQLLLVH
Mflv_1189|M.gilvum_PYR-GCK YVAVPAGQVLPVPDGVPLDHAAGLPEVACTVWSNLVMTARLRAPELLLIH
Mvan_5615|M.vanbaalenii_PYR-1 FVAVPAGQVLPIPDGVPVPHAAGLPEVACTVWSNVVMAAGLTAPELLLVH
MAB_1112c|M.abscessus_ATCC_199 RVNVAATQVLPVPRGVSITAAAALPETAATVWSNVVMTGGLRSGQTLLIH
* *.* *::*:* .* : :*.:**.*.*****:*: . * . :*:*
MSMEG_6362|M.smegmatis_MC2_155 GGASGIGTHAIQVAKAMGARVAVTAGSEEKLGICRDLGADITINYREEDF
TH_1665|M.thermoresistible__bu GGASGVGSHAVQVARALDARVAVTAGTRHKLELCRELGAEVTICYREEDF
Mb3806|M.bovis_AF2122/97 GGASGIGSHAIQVARALAARVAITAGSPEKLELCRDLGAQITINYRDEDF
Rv3777|M.tuberculosis_H37Rv GGASGIGSHAIQVVRALAARVAITAGSPEKLELCRDLGAQITINYRDEDF
MMAR_5332|M.marinum_M GGASGIGSHAIQVCTALGARVAVTAGSSEKLQLCRDLGAQITINYRDEDF
MUL_0096|M.ulcerans_Agy99 GGACGIGSHAIQVCTALGARVAVTAGSSEKLQLSRDLGAQITINYRDEDF
MLBr_00118|M.leprae_Br4923 GGTSGIGSHGIQVARALGAKVAVTAGSEEKLEFCRALGAQITINYRDEDF
MAV_0242|M.avium_104 GGASGIGSHAIQVARALGARVAVTAGSAEKLGFCRELGADITINYRDEDF
Mflv_1189|M.gilvum_PYR-GCK GGASGIGTHAIQVARALNCRVAVTAGSRNKLDLCAELGAEITIDYHHEDF
Mvan_5615|M.vanbaalenii_PYR-1 GGASGIGTHAIQVAKALNCRVAVTAGSRNKLDLCAELGADITIDYHNEDY
MAB_1112c|M.abscessus_ATCC_199 GGGSGIGTHAIQVGRALGANVAVTAGTQFKLDRCRELGAHTLINYREQDF
** .*:*:*.:** *: ..**:***: ** . ***. * *:.:*:
MSMEG_6362|M.smegmatis_MC2_155 VERLRRETD---GADVILDIMGAAYLDRNVDALATGGRLVIIGMQGGVKA
TH_1665|M.thermoresistible__bu VERVRAETDG-AGADVVLDIMGAAYLGRNVDALATDGRLVVIGMQGGRRA
Mb3806|M.bovis_AF2122/97 VARLKQETDG-SGADIILDIMGASYLDRNIDALATDGQLIVIGMQGGVKA
Rv3777|M.tuberculosis_H37Rv VARLKQETDG-SGADIILDIMGASYLDRNIDALATDGQLIVIGMQGGVKA
MMAR_5332|M.marinum_M VARLKQETDG-SGADVILDIMGAAYLDRNVDALAVDGQLVIIGMQGGIKA
MUL_0096|M.ulcerans_Agy99 VARLKQETDG-SGADVILDIMGAAYLDRNVDALAVDGQLVIIGMQGGIKA
MLBr_00118|M.leprae_Br4923 VARLQQQN---GGADVILDIMGAAYLDRNIDALAIDGQLIVIGLQGGVKG
MAV_0242|M.avium_104 VARLADET---GGADVILDIMGAAYLDRNIDALATDGQLVIIGMQGGVKA
Mflv_1189|M.gilvum_PYR-GCK VEVLRGADSDHPGADVILDIMGASYLDRNIDALATGGRLVIIGMQGGTKA
Mvan_5615|M.vanbaalenii_PYR-1 VGVVRDAG----GADVILDIMGAAYLDRNIETLADGGRLVIIGMQGGAKA
MAB_1112c|M.abscessus_ATCC_199 PAVVNAAYS---GANVILDIVGGGYLAKNVEALAENGHLTIIGLQGGAAA
: **:::***:*..** :*:::** .*:* :**:*** .
MSMEG_6362|M.smegmatis_MC2_155 ELNIGKLLGKRAGVIATSLRPRPVDGPGGKGEIVRAVRENVWPMIADGRV
TH_1665|M.thermoresistible__bu ELDLGKLVAKRAAVFGTALRGRPVTGPHGKADIVRAVVENVWPMVASDQV
Mb3806|M.bovis_AF2122/97 ELNLGKLLTKRARVIGTTLRARPVSGPHGKAAIAQAVAASVWPMIAANRV
Rv3777|M.tuberculosis_H37Rv ELNLGKLLTKRARVIGTTLRARPVSGPHGKAAIAQAVAASVWPMIAANRV
MMAR_5332|M.marinum_M ELNLGKLLTKRARVIGTTLRARPLQGPNGKAVIAQAVTASVWPMIASNRV
MUL_0096|M.ulcerans_Agy99 ELNLGKLLTKRARVIGTTLRARPLQGPNGKAVIAQAVTASVWPMIASNRV
MLBr_00118|M.leprae_Br4923 ELNLGKVLSKRARIIGSTLRARPVNGPNSKSEIVAAVTASIWPMIADGSV
MAV_0242|M.avium_104 ELNIGKLLAKRARVIGTTLRGRPVSGPNSKTEIVQAVTASVWPMIADGRV
Mflv_1189|M.gilvum_PYR-GCK EINLGKLLAKRGSVMATALRSRPVTGRGSKSVIVSEVIENVWPLVADGQV
Mvan_5615|M.vanbaalenii_PYR-1 ELNIGKLLTKRGSVIATALRSRPVHGRGGKADIVARVGAELWPLVADGQV
MAB_1112c|M.abscessus_ATCC_199 ELNLGALLFKRGSVHATNLRRRPEQGPGSKAEIIAELRQHLWPLIAEGTV
*:::* :: **. : .: ** ** * .* * : :**::* . *
MSMEG_6362|M.smegmatis_MC2_155 KPIIGAELPIENAQQGHDLLASGKVTGKVLLRVSE-----------
TH_1665|M.thermoresistible__bu RPVVGAEFPIQEAQAAHELLASGDVYGKVLLCVED-----------
Mb3806|M.bovis_AF2122/97 RPVIGTRLPIQQAAQAHELMLSGKTFGKILLTV-------------
Rv3777|M.tuberculosis_H37Rv RPVIGTRLPIQQAAQAHELMLSGKTFGKILLTV-------------
MMAR_5332|M.marinum_M RPIIGSRLPIGQAAQAHQLLLSGQSYGKTLLTVDG-----------
MUL_0096|M.ulcerans_Agy99 RPIIGSRLPIGQAAQAHQLLLSGQSYGKILLTVDG-----------
MLBr_00118|M.leprae_Br4923 RPIIGARMAVQQAADAHQLLLSGKVSGKIVLTVAGSGTETLLSRHH
MAV_0242|M.avium_104 RPIIGARLPIQQAGDAHRRLVASEVTGKIVLTV-------------
Mflv_1189|M.gilvum_PYR-GCK RPVIGAEFPISEAQAAHELLASGDVSGKVILRVDV-----------
Mvan_5615|M.vanbaalenii_PYR-1 RPVIGAEFPIAEAQAAHELLASGDVSGKVLLRVDV-----------
MAB_1112c|M.abscessus_ATCC_199 APVVSAEVPVTDAAEAHRLLDSPETVGKVLLTVRRQ----------
*::.:...: :* .* : : . ** :* *