For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTSTVAAPAQTGTAQSLTLSKLVKTYSSRGRESFTAVKGIDLDIRPGELVALLGPSGCGKTTTLRMIAGL ETVTSGSIKIGDREVSQLPAAKRGVGVGFESYALYPPMSVRENLLYGLKARKVKGAEEMVAAISRRLEMD DMLELRPAGLSSGQKQRVALARALVRNPPVLLLDEPLSHLDASARQRVRRELKVLQREFGYTTIVVTHDQ VEALSLADRLAVMDAGVVQQFGTPDEVFDDPANLFVAQFVGEPQINVVRGTVRVEGGRAHVEIGSRAGAL PTTVTGVADGTPVSVGIRPQDCALRPADATGGVSATVAYFEHLLEFGLATSTVAGVEEGIVVQTPALESY EPEQQVVVTAPPERVYLFDAESGERLR
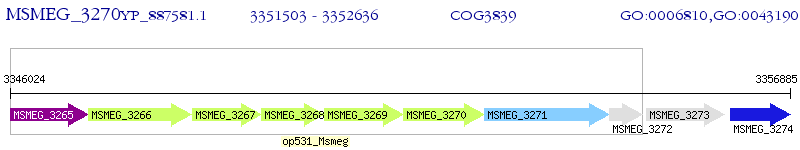
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3270 | - | - | 100% (377) | SN-glycerol-3-phosphate ABC transporter, ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_0518 | - | 3e-74 | 45.38% (346) | sn-glycerol-3-phosphate ABC transporter ATPase |
| M. smegmatis MC2 155 | MSMEG_3108 | - | 1e-70 | 42.90% (366) | ABC transporter, ATPase subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2064c | - | 4e-64 | 38.29% (363) | sugar-transport ATP-binding protein ABC transporter |
| M. gilvum PYR-GCK | Mflv_3778 | - | 7e-68 | 41.32% (363) | ABC transporter related |
| M. tuberculosis H37Rv | Rv2038c | - | 4e-64 | 38.29% (363) | sugar-transport ATP-binding protein ABC transporter |
| M. leprae Br4923 | MLBr_01424 | - | 1e-62 | 42.86% (287) | putative binding-protein-dependent transport protein |
| M. abscessus ATCC 19977 | MAB_1375 | - | 2e-63 | 38.72% (390) | sugar ABC transporter, ATP-binding protein SugC |
| M. marinum M | MMAR_1901 | - | 5e-62 | 38.35% (352) | ABC-type sugar transport protein, ATPase component |
| M. avium 104 | MAV_1377 | sugC | 1e-63 | 37.40% (385) | ABC transporter, ATP-binding protein SugC |
| M. thermoresistible (build 8) | TH_3215 | - | 2e-60 | 42.68% (314) | PUTATIVE ABC transporter ATP-binding protein |
| M. ulcerans Agy99 | MUL_2252 | - | 4e-57 | 37.46% (347) | sugar-transport ATP-binding protein ABC transporter |
| M. vanbaalenii PYR-1 | Mvan_2621 | - | 4e-68 | 42.73% (344) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2064c|M.bovis_AF2122/97 -------------MASVSFEQATRRYPGTDR---PALDRLDLIVGDGEFV
Rv2038c|M.tuberculosis_H37Rv -------------MASVSFEQATRRYPGTDR---PALDRLDLIVGDGEFV
MUL_2252|M.ulcerans_Agy99 -------------MASVSFEGATRRYPGADR---PAVDSLDLVVDDGEFV
MLBr_01424|M.leprae_Br4923 -------------MASVTFEQATRQFPDTDR---PALDRLDLDVDDGEFV
TH_3215|M.thermoresistible__bu -------------MATITYRNASCIYEGSDT---FAVNSLNLEIEDGEFV
MMAR_1901|M.marinum_M -------------MAKVQYSGVTYRYPGSGA---PAVNNLDLEIADGEFL
MAB_1375|M.abscessus_ATCC_1997 -------------MAEIVLNQVSKTYTDG-S---AAVRDVSLNIADGEFI
MAV_1377|M.avium_104 -------------MAEIVLDHVSKSYPDG-A---TAVRDLNLTIADGEFL
Mflv_3778|M.gilvum_PYR-GCK -------------MATVRFSGVTKAYGST-----SVVSDLDLELPDGSLS
Mvan_2621|M.vanbaalenii_PYR-1 -------------MATVRFSGVNKAYGTT-----SVVSDLDLELPDGSFT
MSMEG_3270|M.smegmatis_MC2_155 MTSTVAAPAQTGTAQSLTLSKLVKTYSSRGRESFTAVKGIDLDIRPGELV
: : .: :.* : *.:
Mb2064c|M.bovis_AF2122/97 VLVGPSGCGKTTSLRMVAGLETLDCGRIRIGERDVTEVDPKDRDVAMVFQ
Rv2038c|M.tuberculosis_H37Rv VLVGPSGCGKTTSLRMVAGLETLDCGRIRIGERDVTEVDPKDRDVAMVFQ
MUL_2252|M.ulcerans_Agy99 VLVGPSGCGKTTSLRMLAGLEKVDAGRIRIAERDVTNVDPKSRDVAMVFQ
MLBr_01424|M.leprae_Br4923 VVVGPSGCGKTTSLRMVAGLETLDSGCIRIGDRDVTHVDPKDRDVAMVFQ
TH_3215|M.thermoresistible__bu VLVGPSGSGKSTALRMLAGLEDIDDGDIEIGGRNMRGVPSKDRDIAMVFQ
MMAR_1901|M.marinum_M VLVGPSGCGKSTILRVLAGLESVESGRITIGDTDVTDLPPRARDVAMVFQ
MAB_1375|M.abscessus_ATCC_1997 ILVGPSGCGKSTTLNMIAGLEDISSGELTIAGERVNEKAPKDRDIAMVFQ
MAV_1377|M.avium_104 ILVGPSGCGKTTTLNMIAGLEDISSGELRIGGERVNEKAPKDRDIAMVFQ
Mflv_3778|M.gilvum_PYR-GCK VLVGPSGCGKSTTLRMLAGLETVTSGTITIGDRDVTHLQPRDRDIAMVFQ
Mvan_2621|M.vanbaalenii_PYR-1 VLVGPSGCGKSTSLRMLAGLETVTSGSITIGARDVTHLQPRDRDIAMVFQ
MSMEG_3270|M.smegmatis_MC2_155 ALLGPSGCGKTTTLRMIAGLETVTSGSIKIGDREVSQLPAAKRGVGVGFE
::****.**:* *.::**** : * : *. : . *.:.: *:
Mb2064c|M.bovis_AF2122/97 NYALYPHMTVAQNMGFALKVAKIGKAEIRERVLAAAKLLDLQSYLDRKPK
Rv2038c|M.tuberculosis_H37Rv NYALYPHMTVAQNMGFALKVAKIGKAEIRERVLAAAKLLDLQSYLDRKPK
MUL_2252|M.ulcerans_Agy99 NYALYPHMTVAQNMGFALKVAKTPKAEIRDRVLQAAKLLDLEAYLDRKPK
MLBr_01424|M.leprae_Br4923 NYALYPHMTVAQNLGFALKISKISKAEIHDRILVVAKLLDLQPYLDRKPK
TH_3215|M.thermoresistible__bu NYALYPNKTVAENMGFALKLRGVAASERRRKVEEAAKLLDLTEFLDRKPA
MMAR_1901|M.marinum_M NYALYPNMTVAQNMGFALRNAGMSRADTRQRVLEVASMLELTELLDRKPS
MAB_1375|M.abscessus_ATCC_1997 SYALYPHMTVRQNIAFPLTLAKMSKDDINAKVAEAAKILDLTDYLDRKPA
MAV_1377|M.avium_104 SYALYPHMTVRQNIAFPLTLAKMKKPEIAQKVAETAKILDLTDFLDRKPS
Mflv_3778|M.gilvum_PYR-GCK NYALYPHLTVAENIAFPLRANKTPRREALARAATVAESLGLSALLGRKPK
Mvan_2621|M.vanbaalenii_PYR-1 NYALYPHLTVAENIAFPLRATKTPRREALTRAATIAESLGLAKLLDRKPK
MSMEG_3270|M.smegmatis_MC2_155 SYALYPPMSVRENLLYGLKARKVKGAE--EMVAAISRRLEMDDMLELRPA
.***** :* :*: : * : : * : * :*
Mb2064c|M.bovis_AF2122/97 DLSGGQRQRVAMGRAIVRRPQVFLMDEPLSNLDAKLRGQTRNQIAALQRQ
Rv2038c|M.tuberculosis_H37Rv DLSGGQRQRVAMGRAIVRRPQVFLMDEPLSNLDAKLRGQTRNQIAALQRQ
MUL_2252|M.ulcerans_Agy99 DLSGGQRQRVAMGRAIVRRPQVFLMDEPLSNLDTKLRVQTRNQIAGLQRQ
MLBr_01424|M.leprae_Br4923 DLSGGQRQRVAMGRAIVRRPQVFLMDEPLSNLDAKLRAQTRNQIAELQCQ
TH_3215|M.thermoresistible__bu KLSGGQRQRVAMGRAIVREPQVFCMDEPLSNLDAKLRVQTRTQIAALQRR
MMAR_1901|M.marinum_M KLSGGQRQRVAMGRAIVRRPRVFCMDEPLSNLDAKLRVSTRSHISGLQRQ
MAB_1375|M.abscessus_ATCC_1997 NLSGGQRQRVAMGRAIVRSPKAFLMDEPLSNLDAKLRVQMRTEIARLQKR
MAV_1377|M.avium_104 QLSGGQRQRVAMGRAIVRHPKAFLMDEPLSNLDAKLRVQMRGEIARLQKR
Mflv_3778|M.gilvum_PYR-GCK DLSGGQQQRVAIGRAIIREPSVFLFDEPLSNLDAKLRVETRTELAQIQRR
Mvan_2621|M.vanbaalenii_PYR-1 DLSGGQQQRVAIGRAIIREPSVFLFDEPLSNLDAKLRVETRTELLQIQRR
MSMEG_3270|M.smegmatis_MC2_155 GLSSGQKQRVALARALVRNPPVLLLDEPLSHLDASARQRVRRELKVLQRE
**.**:****:.**::* * .: :*****:**:. * * .: :* .
Mb2064c|M.bovis_AF2122/97 LGTTTVYVTHDQVEAMTMGDRVAVLSDGVLQQCASPRELYRNPGNVFVAG
Rv2038c|M.tuberculosis_H37Rv LGTTTVYVTHDQVEAMTMGDRVAVLSDGVLQQCASPRELYRNPGNVFVAG
MUL_2252|M.ulcerans_Agy99 LGTTTVYVTHDQVEAMTMGDRVAVLCDGVLQQCAAPRELYRRPDNVFVAG
MLBr_01424|M.leprae_Br4923 LGTTTVYVTHDQVEAMTMGDRVAVLCYGVLQQFATPRELYHKPENVFVAG
TH_3215|M.thermoresistible__bu LGTTTVYVTHDQVEAMTMGDRVAVLKHGRLQQFASPGDLYHRPANAFVAG
MMAR_1901|M.marinum_M LGTTTVYVTHDQVEAMTMGHRVAVLKDGVLQQVDTPRELYDNPVNTFVAT
MAB_1375|M.abscessus_ATCC_1997 LGTTTVYVTHDQTEAMTLGDRVVVLRAGKVQQVGAPQELYDTPANLFVAG
MAV_1377|M.avium_104 LGTTTVYVTHDQTEAMTLGDRVVVMHSGVAQQIGTPDELYENPANLFVAG
Mflv_3778|M.gilvum_PYR-GCK LGITSVYVTHDQEEAMTLSDRMIVMRDGKIAQQGAPQDVYARPADTFVAA
Mvan_2621|M.vanbaalenii_PYR-1 LGITSVYVTHDQEEAMTLSDRMVVMRDGRIAQQGTPQEVYARPADTFVAA
MSMEG_3270|M.smegmatis_MC2_155 FGYTTIVVTHDQVEALSLADRLAVMDAGVVQQFGTPDEVFDDPANLFVAQ
:* *:: ***** **:::..*: *: * * :* ::: * : ***
Mb2064c|M.bovis_AF2122/97 FIGSPAMNLFRLSIADST----VSLGDWQILLPRAVVGTAAE------VI
Rv2038c|M.tuberculosis_H37Rv FIGSPAMNLFRLSIADST----VSLGDWQILLPRAVVGTAAE------VI
MUL_2252|M.ulcerans_Agy99 FIGSPAMNMFILPIVDSS----VLLGDWLIQLPREVTVPAPE------VV
MLBr_01424|M.leprae_Br4923 FIGSPGMNLVTLSIVDSS----VLLGDWPIRIPREIASAASE------VI
TH_3215|M.thermoresistible__bu FIGSPAMNLFNAQLVPEG----VQIGDTVLELERAQLTALHEAK-LDTVT
MMAR_1901|M.marinum_M FIGAPAMNLIEAPVADGV----ARLSGLVIPVPRGAADR---------VL
MAB_1375|M.abscessus_ATCC_1997 FIGSPSMNFFPATLTDVG----VQLPFGEVTLDAGLYAGITARKPSGDVI
MAV_1377|M.avium_104 FIGSPAMNFFPATLTPIG----LKVPFGEVMLTPEVQQVIAEHPEPDNVI
Mflv_3778|M.gilvum_PYR-GCK FVGSPKMNLLEGTLTGGT----FTLANG-FALPIDEGATDGP------LT
Mvan_2621|M.vanbaalenii_PYR-1 FVGSPKMNLIDGAVSGTT----FTVGDG-FALTVDQTAAGGP------VT
MSMEG_3270|M.smegmatis_MC2_155 FVGEPQINVVRGTVRVEGGRAHVEIGSRAGALPTTVTGVADGTP----VS
*:* * :*.. : : : :
Mb2064c|M.bovis_AF2122/97 IGVRPEHLELG---GAG-------IEMDVDMVEELGADAYLYGR------
Rv2038c|M.tuberculosis_H37Rv IGVRPEHLELG---GAG-------IEMDVDMVEELGADAYLYGR------
MUL_2252|M.ulcerans_Agy99 VGVRPEHFEVG---NLG-------VEMEIDVVEELGADAYLYGR------
MLBr_01424|M.leprae_Br4923 IGVRPEHFELG---SLG-------VEVEIDMVEELGADAYLYGR------
TH_3215|M.thermoresistible__bu VGLRPEHLELS---DAGG------VEVVVDLVEDLGSEAYLYTH------
MMAR_1901|M.marinum_M IGVRPESWDVVGGDAAAA------LGVRVEQVEELGFESFIYATPV----
MAB_1375|M.abscessus_ATCC_1997 VGIRPEQFEDAALVDTYKRITGLTLTVNADVVESLGSDKYVHFTTEGGAA
MAV_1377|M.avium_104 VGARPEHLSDAALIDGYQRIRALTFEVKVDMVESLGADKYVYFSTAAWAA
Mflv_3778|M.gilvum_PYR-GCK LGVRPDDLLPTVGAADA--------PARVALIEHLGPRAIVTVD------
Mvan_2621|M.vanbaalenii_PYR-1 LGVRPDDLLPTVSDGDG--------AARVILIEHLGPRAIVTID------
MSMEG_3270|M.smegmatis_MC2_155 VGIRPQDCALRPADATGG------VSATVAYFEHLLEFGLATST------
:* **: .* *
Mb2064c|M.bovis_AF2122/97 -------IVSGGCEMDQSIVARVDGRGPPERGSRVRLCP-TPGHLHFFAV
Rv2038c|M.tuberculosis_H37Rv -------IVSGGCEMDQSIVARVDGRGPPERGSRVRLCP-TPGHLHFFAV
MUL_2252|M.ulcerans_Agy99 -------ISNGGTMIDQSVVARADGSNPPERGSRVRLYP-QPAQLHFFTV
MLBr_01424|M.leprae_Br4923 -------IAGASKVTDQLVVARVDGRNPPAKGSRVRLYT-EPGNVHFFGV
TH_3215|M.thermoresistible__bu -------TGSG-----VDLVARCLDRVPAKLADTVRLRKRTDGVVHLFHP
MMAR_1901|M.marinum_M -------AQDGWSSRTRRIVIRSDRHTTVGAGDSLSIAP-NPQEVCFFDS
MAB_1375|M.abscessus_ATCC_1997 HSDELTELAQESEVAENEFVARLSAASKVAEGQPIELII-DTGKLVIFDA
MAV_1377|M.avium_104 HSTQLDELAAEADAHENQFVARVPAESKAAIGQTVELAL-DTTKLMVFDA
Mflv_3778|M.gilvum_PYR-GCK --------ARGTELTSVVDTARLSG---ITEGTGVELAV-RQGATHLFDT
Mvan_2621|M.vanbaalenii_PYR-1 --------ARGTELTSVVETSRLSG---ITEGTPVDLAV-RPGATHFFDP
MSMEG_3270|M.smegmatis_MC2_155 -----------VAGVEEGIVVQTPALESYEPEQQVVVTA-PPERVYLFDA
. : : : .*
Mb2064c|M.bovis_AF2122/97 D-GRRIPG------
Rv2038c|M.tuberculosis_H37Rv D-GRRIPG------
MUL_2252|M.ulcerans_Agy99 D-GRRIA-------
MLBr_01424|M.leprae_Br4923 D-GHRIS-------
TH_3215|M.thermoresistible__bu DTGERVGE------
MMAR_1901|M.marinum_M RTESRIR-------
MAB_1375|M.abscessus_ATCC_1997 ESGENLSLAATAE-
MAV_1377|M.avium_104 DSGVNLTVAPSDSP
Mflv_3778|M.gilvum_PYR-GCK TTGRRIAGETR---
Mvan_2621|M.vanbaalenii_PYR-1 ATGSRLSG------
MSMEG_3270|M.smegmatis_MC2_155 ESGERLR-------
.: