For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LAEVEFRDVTRSYPGGVQALKGLDLTVADGEFLILVGPSGCGKSTALRMLAGLDKPTAGEIRIGGAVVNH LSPGQRDIAMVFQNYALYPHMSVYRNLAYGLRQRRTPRAEIDRRVRETAELLQITEFLDRKPGQLSGGQR QRVAMGRALVREPQAFLLDEPLSNLDAKLRNQVRGDLKRLHREVPVTSIYVTHDQVEAMTLGDRLCVMSQ GEVQQIGTTDDIYNRPANTFVAAFMGSPPMNLMPGIVRQGVLHVGGESVTPVSCPEGPVTVGARPEHLEL SPVWGDGMVPARVDFVEPLGSHSLVTALLGDDPASTTRVIVQAPADTDLPSGTRIGLTLPPQRTYFFDAE SGDALTAASMGRERISLA
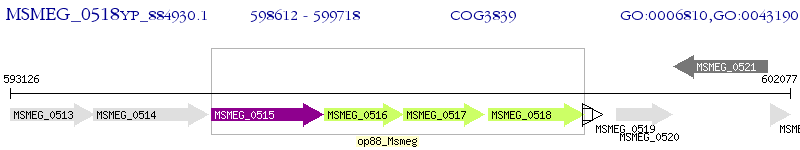
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0518 | - | - | 100% (368) | sn-glycerol-3-phosphate ABC transporter ATPase |
| M. smegmatis MC2 155 | MSMEG_5058 | - | 2e-94 | 50.51% (392) | ABC transporter, ATP-binding protein SugC |
| M. smegmatis MC2 155 | MSMEG_3108 | - | 7e-86 | 48.75% (359) | ABC transporter, ATPase subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1270 | sugC | 2e-88 | 46.39% (388) | sugar-transport ATP-binding protein ABC transporter SugC |
| M. gilvum PYR-GCK | Mflv_3778 | - | 3e-87 | 48.02% (354) | ABC transporter related |
| M. tuberculosis H37Rv | Rv1238 | sugC | 2e-88 | 46.39% (388) | sugar-transport ATP-binding protein ABC transporter SugC |
| M. leprae Br4923 | MLBr_01089 | - | 4e-90 | 47.81% (389) | putative ABC-transport protein, ATP-binding component |
| M. abscessus ATCC 19977 | MAB_1375 | - | 4e-85 | 44.33% (388) | sugar ABC transporter, ATP-binding protein SugC |
| M. marinum M | MMAR_4203 | sugC | 4e-88 | 45.88% (388) | sugar-transport ATP-binding protein ABC transporter SugC |
| M. avium 104 | MAV_1377 | sugC | 5e-94 | 49.35% (387) | ABC transporter, ATP-binding protein SugC |
| M. thermoresistible (build 8) | TH_4034 | - | 6e-91 | 48.07% (389) | PUTATIVE ABC transporter, ATP-binding protein SugC |
| M. ulcerans Agy99 | MUL_2252 | - | 1e-85 | 48.45% (355) | sugar-transport ATP-binding protein ABC transporter |
| M. vanbaalenii PYR-1 | Mvan_2621 | - | 3e-89 | 49.44% (358) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1270|M.bovis_AF2122/97 MAEIVLDHVNKSYPDGH----TAVRDLNLTIADGEFLILVGPSGCGKTTT
Rv1238|M.tuberculosis_H37Rv MAEIVLDHVNKSYPDGH----TAVRDLNLTIADGEFLILVGPSGCGKTTT
MMAR_4203|M.marinum_M MAEIVLEHVNKSYPDGH----TAVRDLNITISDGEFLILVGPSGCGKTTT
MAV_1377|M.avium_104 MAEIVLDHVSKSYPDGA----TAVRDLNLTIADGEFLILVGPSGCGKTTT
MLBr_01089|M.leprae_Br4923 MAEIVLEHVNKNYPNGA----TAVHDLSITVADGEFLILIGPSGCGKTTT
MAB_1375|M.abscessus_ATCC_1997 MAEIVLNQVSKTYTDGS----AAVRDVSLNIADGEFIILVGPSGCGKSTT
TH_4034|M.thermoresistible__bu MAEIVLDKVTKSYPDGAGGVRHAVKDLSLTIADGEFIILVGPSGCGKSTT
MUL_2252|M.ulcerans_Agy99 MASVSFEGATRRYPGADR---PAVDSLDLVVDDGEFVVLVGPSGCGKTTS
Mflv_3778|M.gilvum_PYR-GCK MATVRFSGVTKAY-GSTS----VVSDLDLELPDGSLSVLVGPSGCGKSTT
Mvan_2621|M.vanbaalenii_PYR-1 MATVRFSGVNKAY-GTTS----VVSDLDLELPDGSFTVLVGPSGCGKSTS
MSMEG_0518|M.smegmatis_MC2_155 MAEVEFRDVTRSYPGGVQ----ALKGLDLTVADGEFLILVGPSGCGKSTA
** : : ..: * . .: .:.: : **.: :*:*******:*:
Mb1270|M.bovis_AF2122/97 LNMIAGLEDISSGELRIAGERVNEKAPKDRDIAMVFQSYALYPHMTVRQN
Rv1238|M.tuberculosis_H37Rv LNMIAGLEDISSGELRIAGERVNEKAPKDRDIAMVFQSYALYPHMTVRQN
MMAR_4203|M.marinum_M LNMIAGLEDISSGELRIGGERVNEKAPKDRDIAMVFQSYALYPHMTVRQN
MAV_1377|M.avium_104 LNMIAGLEDISSGELRIGGERVNEKAPKDRDIAMVFQSYALYPHMTVRQN
MLBr_01089|M.leprae_Br4923 LNMIAGLEDISSGELRIDGDRVNEKAPKDRDIAMVFQSYALYPHMTVRQN
MAB_1375|M.abscessus_ATCC_1997 LNMIAGLEDISSGELTIAGERVNEKAPKDRDIAMVFQSYALYPHMTVRQN
TH_4034|M.thermoresistible__bu LNMIAGLEDITSGEIRIDGERVNEVAPKDRDIAMVFQSYALYPHMTVRQN
MUL_2252|M.ulcerans_Agy99 LRMLAGLEKVDAGRIRIAERDVTNVDPKSRDVAMVFQNYALYPHMTVAQN
Mflv_3778|M.gilvum_PYR-GCK LRMLAGLETVTSGTITIGDRDVTHLQPRDRDIAMVFQNYALYPHLTVAEN
Mvan_2621|M.vanbaalenii_PYR-1 LRMLAGLETVTSGSITIGARDVTHLQPRDRDIAMVFQNYALYPHLTVAEN
MSMEG_0518|M.smegmatis_MC2_155 LRMLAGLDKPTAGEIRIGGAVVNHLSPGQRDIAMVFQNYALYPHMSVYRN
*.*:***: :* : * *.. * .**:*****.******::* .*
Mb1270|M.bovis_AF2122/97 IAFPLTLAKMRKADIAQKVSETAKILDLTNLLDRKPSQLSGGQRQRVAMG
Rv1238|M.tuberculosis_H37Rv IAFPLTLAKMRKADIAQKVSETAKILDLTNLLDRKPSQLSGGQRQRVAMG
MMAR_4203|M.marinum_M IAFPLTLAKMKKAAIAQKVEETAKILDLTSFLDRKPSQLSGGQRQRVAMG
MAV_1377|M.avium_104 IAFPLTLAKMKKPEIAQKVAETAKILDLTDFLDRKPSQLSGGQRQRVAMG
MLBr_01089|M.leprae_Br4923 IAFPLMLAKVKKAEIAQKVSETAQILDLTDLLDRKPSQLSGGQRQRVAMG
MAB_1375|M.abscessus_ATCC_1997 IAFPLTLAKMSKDDINAKVAEAAKILDLTDYLDRKPANLSGGQRQRVAMG
TH_4034|M.thermoresistible__bu IAFPLTLAKMKKADIARKVEETAKILDLTELLDRKPAQLSGGQRQRVAMG
MUL_2252|M.ulcerans_Agy99 MGFALKVAKTPKAEIRDRVLQAAKLLDLEAYLDRKPKDLSGGQRQRVAMG
Mflv_3778|M.gilvum_PYR-GCK IAFPLRANKTPRREALARAATVAESLGLSALLGRKPKDLSGGQQQRVAIG
Mvan_2621|M.vanbaalenii_PYR-1 IAFPLRATKTPRREALTRAATIAESLGLAKLLDRKPKDLSGGQQQRVAIG
MSMEG_0518|M.smegmatis_MC2_155 LAYGLRQRRTPRAEIDRRVRETAELLQITEFLDRKPGQLSGGQRQRVAMG
:.: * : : :. *: * : *.*** :*****:****:*
Mb1270|M.bovis_AF2122/97 RAIVRHPKAFLMDEPLSNLDAKLRVQMRGEIAQLQRRLGTTTVYVTHDQT
Rv1238|M.tuberculosis_H37Rv RAIVRHPKAFLMDEPLSNLDAKLRVQMRGEIAQLQRRLGTTTVYVTHDQT
MMAR_4203|M.marinum_M RAIVRHPKAFLMDEPLSNLDAKLRVQMRGEIARLQKRLGTTTVYVTHDQT
MAV_1377|M.avium_104 RAIVRHPKAFLMDEPLSNLDAKLRVQMRGEIARLQKRLGTTTVYVTHDQT
MLBr_01089|M.leprae_Br4923 RAIVRHPKAFLMDEPLSNLDAKLRVRTRGEIARLQRRLGATTVYVTHDQT
MAB_1375|M.abscessus_ATCC_1997 RAIVRSPKAFLMDEPLSNLDAKLRVQMRTEIARLQKRLGTTTVYVTHDQT
TH_4034|M.thermoresistible__bu RAIVRQPKAFLMDEPLSNLDAKLRVQMRSEIARLQSRLRTTTVYVTHDQT
MUL_2252|M.ulcerans_Agy99 RAIVRRPQVFLMDEPLSNLDTKLRVQTRNQIAGLQRQLGTTTVYVTHDQV
Mflv_3778|M.gilvum_PYR-GCK RAIIREPSVFLFDEPLSNLDAKLRVETRTELAQIQRRLGITSVYVTHDQE
Mvan_2621|M.vanbaalenii_PYR-1 RAIIREPSVFLFDEPLSNLDAKLRVETRTELLQIQRRLGITSVYVTHDQE
MSMEG_0518|M.smegmatis_MC2_155 RALVREPQAFLLDEPLSNLDAKLRNQVRGDLKRLHREVPVTSIYVTHDQV
**::* *..**:********:*** . * :: :: .: *::******
Mb1270|M.bovis_AF2122/97 EAMTLGDRVVVMYGGIAQQIGTPEELYERPANLFVAGFIGSPAMNFFPAR
Rv1238|M.tuberculosis_H37Rv EAMTLGDRVVVMYGGIAQQIGTPEELYERPANLFVAGFIGSPAMNFFPAR
MMAR_4203|M.marinum_M EAMTLGDRVVVMHAGAAQQIGTPQELYERPANLFVAGFIGSPAMNFFPAV
MAV_1377|M.avium_104 EAMTLGDRVVVMHSGVAQQIGTPDELYENPANLFVAGFIGSPAMNFFPAT
MLBr_01089|M.leprae_Br4923 EAMTLGDRVVVMRSGVVQQIGTPDELYERPVNLFVAGFIGSPVMNFFPAK
MAB_1375|M.abscessus_ATCC_1997 EAMTLGDRVVVLRAGKVQQVGAPQELYDTPANLFVAGFIGSPSMNFFPAT
TH_4034|M.thermoresistible__bu EAMTLGDRVVVMLDGVAQQIGTPDELYTQPANLFVAGFIGSPGMNFFPAS
MUL_2252|M.ulcerans_Agy99 EAMTMGDRVAVLCDGVLQQCAAPRELYRRPDNVFVAGFIGSPAMNMFILP
Mflv_3778|M.gilvum_PYR-GCK EAMTLSDRMIVMRDGKIAQQGAPQDVYARPADTFVAAFVGSPKMNLLEGT
Mvan_2621|M.vanbaalenii_PYR-1 EAMTLSDRMVVMRDGRIAQQGTPQEVYARPADTFVAAFVGSPKMNLIDGA
MSMEG_0518|M.smegmatis_MC2_155 EAMTLGDRLCVMSQGEVQQIGTTDDIYNRPANTFVAAFMGSPPMNLMPGI
****:.**: *: * * .:. ::* * : ***.*:*** **::
Mb1270|M.bovis_AF2122/97 LTAIGLTLPFGEVTLAPEVQGVIAAHPKPENVIVGVRPEHIQDAALIDAY
Rv1238|M.tuberculosis_H37Rv LTAIGLTLPFGEVTLAPEVQGVIAAHPKPENVIVGVRPEHIQDAALIDAY
MMAR_4203|M.marinum_M LTPTGLTLPFGEVTLDPQVQQVIAQHPRPANIIVGIRPEQIQDAALIDAY
MAV_1377|M.avium_104 LTPIGLKVPFGEVMLTPEVQQVIAEHPEPDNVIVGARPEHLSDAALIDGY
MLBr_01089|M.leprae_Br4923 LTPNGLTLPLGELNLSRDVQAMIGQHPVPDRVIVGVRPEHLTDAKLIDAH
MAB_1375|M.abscessus_ATCC_1997 LTDVGVQLPFGEVTLDAGLYAGITARKPSGDVIVGIRPEQFEDAALVDTY
TH_4034|M.thermoresistible__bu LTDAGVRLPFGEALLPPDVLDTVRRHEKTDNIIAGLRPEHLEDVSLLDTY
MUL_2252|M.ulcerans_Agy99 IVDSSVLLGDWLIQLPREVT------VPAPEVVVGVRPEHFE--------
Mflv_3778|M.gilvum_PYR-GCK LTGG-------TFTLANGFALPIDEGATDGPLTLGVRPD--DLLPT----
Mvan_2621|M.vanbaalenii_PYR-1 VSGT-------TFTVGDGFALTVDQTAAGGPVTLGVRPD--DLLPT----
MSMEG_0518|M.smegmatis_MC2_155 VRQG-------VLHVGGESVTPVS--CPEGPVTVGARPEHLELSPV----
: : : * **:
Mb1270|M.bovis_AF2122/97 QRIRALTFQVKVNLVESLGADKYLYFTTESPAVHSVQLDELAEVEGESAL
Rv1238|M.tuberculosis_H37Rv QRIRALTFQVKVNLVESLGADKYLYFTTESPAVHSVQLDELAEVEGESAL
MMAR_4203|M.marinum_M QRIRALTFEVNTDLVESLGSDKYVYFSTAGCDVHSAQLDELAGLEGESEV
MAV_1377|M.avium_104 QRIRALTFEVKVDMVESLGADKYVYFSTAAWAAHSTQLDELA---AEADA
MLBr_01089|M.leprae_Br4923 QHGTVLTFKVKVDLVESLGADKYIYFTTTGCDVYSAQLDELA---SELEV
MAB_1375|M.abscessus_ATCC_1997 KRITGLTLTVNADVVESLGSDKYVHFTTEGGAAHSDELTELAQ---ESEV
TH_4034|M.thermoresistible__bu ARLGAVTFDVNVELVESLGAEKYVHFRAEGAGAHAAQLAQLA---AESGA
MUL_2252|M.ulcerans_Agy99 --VGNLGVEMEIDVVEELGADAYLYGRISNGGT----------------M
Mflv_3778|M.gilvum_PYR-GCK --VGAADAPARVALIEHLGPRAIVT--------------------VDARG
Mvan_2621|M.vanbaalenii_PYR-1 --VSDGDGAARVILIEHLGPRAIVT--------------------IDARG
MSMEG_0518|M.smegmatis_MC2_155 --WGDGMVPARVDFVEPLGSHSLVTALL-----------------GDDPA
. .:* **. :
Mb1270|M.bovis_AF2122/97 HENQFVARVPAESKVAIGQSVELAFDTARLAVFDADSGANLTIPHRA---
Rv1238|M.tuberculosis_H37Rv HENQFVARVPAESKVAIGQSVELAFDTARLAVFDADSGANLTIPHRA---
MMAR_4203|M.marinum_M IENRFVARVSAESRATLGRPIELAFDTTRLTVFDADTGANLTNPIAAVQ-
MAV_1377|M.avium_104 HENQFVARVPAESKAAIGQTVELALDTTKLMVFDADSGVNLTVAPSDSP-
MLBr_01089|M.leprae_Br4923 RENQFVARVSAESKMAIGESIELAFGTAKIAVFDADSGVNLTVSAPTDA-
MAB_1375|M.abscessus_ATCC_1997 AENEFVARLSAASKVAEGQPIELIIDTGKLVIFDAESGENLSLAATAE--
TH_4034|M.thermoresistible__bu GENEFIARVSTESKAKKGQSLQLAFNPAKLLVFDADTGANLTLPPSDA--
MUL_2252|M.ulcerans_Agy99 IDQSVVARADGSNPPERGSRVRLYPQPAQLHFFTVD-GRRIA--------
Mflv_3778|M.gilvum_PYR-GCK TELTSVVDTARLSGITEGTGVELAVRQGATHLFDTTTGRRIAGETR----
Mvan_2621|M.vanbaalenii_PYR-1 TELTSVVETSRLSGITEGTPVDLAVRPGATHFFDPATGSRLSG-------
MSMEG_0518|M.smegmatis_MC2_155 STTRVIVQAPADTDLPSGTRIGLTLPPQRTYFFDAESGDALTAASMGRER
:. . * : * .* * ::
Mb1270|M.bovis_AF2122/97 ----
Rv1238|M.tuberculosis_H37Rv ----
MMAR_4203|M.marinum_M ----
MAV_1377|M.avium_104 ----
MLBr_01089|M.leprae_Br4923 ----
MAB_1375|M.abscessus_ATCC_1997 ----
TH_4034|M.thermoresistible__bu ----
MUL_2252|M.ulcerans_Agy99 ----
Mflv_3778|M.gilvum_PYR-GCK ----
Mvan_2621|M.vanbaalenii_PYR-1 ----
MSMEG_0518|M.smegmatis_MC2_155 ISLA