For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MITRFIRTQLIIFTITSVVASAMMVFSYLQAPVLFGIGRITVTLELPSTGGLYRFSNVTYRGIEVGKVTD VRPTRSGALATLSLQTSPRIPADLQADVRSVSAVGEQYVDLRPRTDDAPYLQDGSVITAANTSVPQKVGP MLDQMSALVDSIPSGQLGGLLDESFRALNGSGHELGALFDSSARLASDLDATADTTRTLIDDSRPLLDGQ AASVDAIRTWARSLAGITRQVADDDAQIRTIVQTGSGAADEASRLLDQVKPTLPVLLANLVTVGQIGVTY NPSIEQLLVLLPPYLAATQSYGSSLNNPTGMALSEFSLTLGDPPACTVGFLPPSEWRSPADLTDVDTPDG LYCKLPQDSPIGVRGARNFPCMGQPGKRAPTVEICESDRPFEPLAMRQHVTGPYPIDPNLIAQGVPPDDR VTLDEHIYGPVEGTPVDGDALPVAPSAHTAGDGPSVAIATYDPETGRYATPDGQVHHQADLATPGAQNWE DLLPR
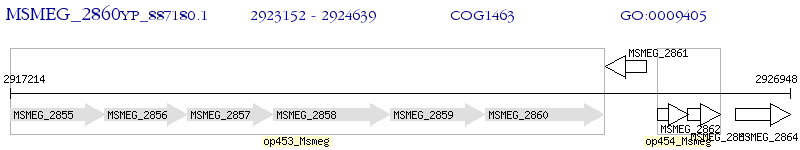
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2860 | - | - | 100% (495) | virulence factor mce family protein |
| M. smegmatis MC2 155 | MSMEG_4785 | - | e-173 | 65.05% (455) | mce-family protein mce1f |
| M. smegmatis MC2 155 | MSMEG_0351 | - | e-102 | 43.17% (505) | virulence factor mce family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0180 | mce1F | 4e-92 | 38.17% (524) | MCE-family protein MCE1F |
| M. gilvum PYR-GCK | Mflv_4533 | - | 0.0 | 75.99% (504) | virulence factor Mce family protein |
| M. tuberculosis H37Rv | Rv1971 | mce3F | 4e-91 | 44.52% (420) | MCE-family protein MCE3F |
| M. leprae Br4923 | MLBr_02594 | mce1F | 3e-85 | 35.44% (522) | putative secreted protein |
| M. abscessus ATCC 19977 | MAB_4148c | - | 3e-94 | 39.57% (508) | MCE-family protein |
| M. marinum M | MMAR_4705 | - | 0.0 | 60.41% (543) | MCE-family protein |
| M. avium 104 | MAV_0953 | - | 1e-174 | 64.54% (454) | mce-family protein mce1f |
| M. thermoresistible (build 8) | TH_4161 | - | 0.0 | 77.45% (501) | PUTATIVE virulence factor mce family protein |
| M. ulcerans Agy99 | MUL_3923 | mce3F_1 | 1e-104 | 42.74% (503) | MCE-family protein Mce3F_1 |
| M. vanbaalenii PYR-1 | Mvan_4145 | - | 1e-167 | 53.39% (560) | virulence factor Mce family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2860|M.smegmatis_MC2_155 --MITRFIRTQLIIFTITSVVASAMMVFSYLQAPVLFG-IGRITVTLELP
TH_4161|M.thermoresistible__bu --MLTRFVRTQLVIFTIAAVVGMGAMVFGYLQVPMLLG-IGRISVTMELP
Mflv_4533|M.gilvum_PYR-GCK --MLTRFVRTQLIIFSIAAVVGLGSMVFVYLQAPVMLG-IGRMTVTLKLP
MMAR_4705|M.marinum_M --MLTRFVRIQLAIFAIVGMVGVAVMAVWYIQAPTLLG-IGKMTVTLELP
MAV_0953|M.avium_104 --MLTRFVRIQLAIFLIAGTIGVISMVLFYIQAPTLLG-IGRMTVTLELP
Mvan_4145|M.vanbaalenii_PYR-1 --MLTRFVRNQLIIFTIASIVGVAVMLFAYMQVPTLLG-VGRLVVTLELP
MAB_4148c|M.abscessus_ATCC_199 ---MERLIKIQLAIFAAVTVISVALMAIFYLRVPTALG-IGKHDVKAQFV
Rv1971|M.tuberculosis_H37Rv MLHLPRRVIVQLAVFTVIAVGVLAITFLHFVRLPAMLFGVGRYTVTMELV
MUL_3923|M.ulcerans_Agy99 -MRLTRQIFIQMAIFAIVATTALLIMVFGYMRVPAMVG-IGQYRVTVELP
Mb0180|M.bovis_AF2122/97 --MLTRFIRRQLILFAIVSVVAIVVLGWYYLRIPSLVG-IGQYTLKADLP
MLBr_02594|M.leprae_Br4923 --MLTPLIRRQLVLFAILTVISLTVLGVYYLQIPSLVG-IGRYSLKAELS
: : *: :* ::: * . :*: :. .:
MSMEG_2860|M.smegmatis_MC2_155 STGGLYRFSNVTYRGIEVGKVTDVRPTRSGALATLSLQTSPRIPADLQAD
TH_4161|M.thermoresistible__bu STGGLYRFSNVTYRGVKVGKVLDVRPTRTGAVVTMSLQSRPRIPADLQAD
Mflv_4533|M.gilvum_PYR-GCK NTGGLYEFSNVTYRGIEVGKVTGIEPTGDGVTVTMSLKSSPKIPTDLKAH
MMAR_4705|M.marinum_M ATGGLYRFSNVTYRGVQIGKVTALGLTSSGAKATLALDASPKIPADLKAE
MAV_0953|M.avium_104 ATGGLYRFSNVTYRGVQLGKVTAVGLTPSGVKATLSLSTSPKVPADLTAE
Mvan_4145|M.vanbaalenii_PYR-1 ESGGLYRFSNVTYRGVQVGKVTGVSLTENGAEATLSLETSPKIPADLQAE
MAB_4148c|M.abscessus_ATCC_199 ASGGLYQNANVTFRGVQIGRVTDVELTPEGVTATMRLNDNVKVPGNVVAA
Rv1971|M.tuberculosis_H37Rv EAGGLYRTGNVTYRGFEVGRVAAVRLTDTGVQAVLALKSGIDIPSDLKAE
MUL_3923|M.ulcerans_Agy99 EAGGLYPRSNVTYRGVEVGTVKSVHLTNTGVAAVLSLNSDVKIPADVEAE
Mb0180|M.bovis_AF2122/97 ASGGLYPTANVTYRGITIGKVTAVEPTDQGARVTMSIASNYKIPVDASAN
MLBr_02594|M.leprae_Br4923 ATGGLYPTANVTYRGIAIGKVTDVEPTATGAEATMSIDSRYKIPIDSAAN
:**** .***:**. :* * : * *. ..: : :* : *
MSMEG_2860|M.smegmatis_MC2_155 VRSVSAVGEQYVDLRPR-TDDAPYLQDGSVITAANTSVPQKVGPMLDQMS
TH_4161|M.thermoresistible__bu VRSVSAVGEQYVELLPR-HDGAPYLEDGSVIGIENTSVPQRVGPMLDQMS
Mflv_4533|M.gilvum_PYR-GCK VRSVSAVGEQYVDLEPR-SDAGPYLQDGSVITGDRSSIPQAVGPMLDQMS
MMAR_4705|M.marinum_M VRSISAVGEYYVDLRPR-TDSPPYLHDGSVIARDRAVIGQPIGPVLDQTS
MAV_0953|M.avium_104 VLSVSAVGEQYVDLRPR-TNSPPYLHDGSVIAMRDTKIPQPVGPMLDQVN
Mvan_4145|M.vanbaalenii_PYR-1 VLSVSAVGEQYVDLRPN-TDEGPYLEDGARIPVANTTVPQQVGPMLDQLS
MAB_4148c|M.abscessus_ATCC_199 VKSVSAIGEQYVDLIPPPTGAAGTLRSGDVIGADRTSIPTDVSTLLRQAD
Rv1971|M.tuberculosis_H37Rv VHSHTAIGETYVELLPR-NAASPPLKNGDVIALADTSVPPDINDLLSAAN
MUL_3923|M.ulcerans_Agy99 VHSVSSVGEQYVQLLPR-SGQGPSLKDGDVIPSSRTRVPTDINTILDETN
Mb0180|M.bovis_AF2122/97 VHSVSAVGEQYIDLVST-GAPGKYFSSGQTIT--KGTVPSEIGPALDNSN
MLBr_02594|M.leprae_Br4923 VHSVSAVGEQYLDLVSS-GNPGKYLAPGQTIT--KSKIPIEIGPALDSVN
* * :::** *::* . : * * : :. * .
MSMEG_2860|M.smegmatis_MC2_155 ALVDSIPSGQLGGLLDESFRALNGSGHELGALFDSSARLASDLDATADTT
TH_4161|M.thermoresistible__bu ALIDSIPSGQLGALLDESATALGGSGYELGSAFDSAAQLAERLNGSGERA
Mflv_4533|M.gilvum_PYR-GCK ALVDSIPTGTLSGLLDESYAALNGSGYELGSLFDSSARLAADADSTGDRM
MMAR_4705|M.marinum_M ALINSIPKGKLSTLLDESFNAFNGAGYDFGSLFDSSAQLSGDLNGIADRT
MAV_0953|M.avium_104 ALVQSIPKTKLGQLLDESFQGFNGSGYDLGSLIDSSQTLSRDANGVVDRT
Mvan_4145|M.vanbaalenii_PYR-1 ALVGSLPKDRIPDLLDETFKAFNGAGPDFGSMLDSAATLTNDVNGVSDQM
MAB_4148c|M.abscessus_ATCC_199 GLVNTVADTRLREVLAEAFKAFNGSGQELSRLIDSSRLLIQEANTNYDST
Rv1971|M.tuberculosis_H37Rv TALEAIPHENLQTVIDESYTAVAGLGLELSRLIKGSAELAIDARANLDPL
MUL_3923|M.ulcerans_Agy99 RGLQAIPRENLKTVVDEAAVAVGGLGPELRRLITGSSKLAIDARANLDEL
Mb0180|M.bovis_AF2122/97 RGLAALPTEKIGLLLDETAQAVGGLGPALQRLVDSTQAIVGDFKTNIGDV
MLBr_02594|M.leprae_Br4923 RGLAALPKEKIGSLLDETAQAVGGLGPALQRLVSSTQAIVDAFKTNITDV
: ::. : :: *: .. * * : . .: :
MSMEG_2860|M.smegmatis_MC2_155 RTLIDDSRPLLDGQAASVDAIRTWARSLAGITRQVADDDAQIRTIVQTGS
TH_4161|M.thermoresistible__bu RVLIEDGRPLLDGQAVGAEAIRTWARSLSGITGQLAADDAQIRTILTDGP
Mflv_4533|M.gilvum_PYR-GCK RNLITDSEPLLDGQAQKADAITTWARSLAGITRQIADDDQAVRTILQTGP
MMAR_4705|M.marinum_M RTLTEDSGPFLDAQAQTTDSIRLWARSLAGVSGQLATNDPQIRTLLERGP
MAV_0953|M.avium_104 RALTEDTGPLLDTQAQTTDSIRTWARSFAGISDVMVNNDSHFRTILEKGP
Mvan_4145|M.vanbaalenii_PYR-1 RSLIDDSGPLIDSQAETADAIRTWAQSLNGISAQVVQNDPQVRALLQQGP
MAB_4148c|M.abscessus_ATCC_199 TKLIDQVGPLLDAQIRSGDNIRGSVSSLAALTEHFAKADPQLRQFLKVAP
Rv1971|M.tuberculosis_H37Rv VALIDRAGPVLDSQTHTSDAIAAWAAQLAAVTGQLQTHDSAVGDLIDRGG
MUL_3923|M.ulcerans_Agy99 TLLIDQSKPVLDTQTDTAGAIRSWASNLASITDQLQRQDSSLAGLLDQGP
Mb0180|M.bovis_AF2122/97 NDIIENSGPILDSQVNTGDQIERWARKLNNLAAQTATRDQNVRSILSQAA
MLBr_02594|M.leprae_Br4923 NDIIQNSGPILDSQVKSDNAIERWVHNLNVLAAQSARQDQHLKSILSQAA
: *.:* * * . .: :: * . :: .
MSMEG_2860|M.smegmatis_MC2_155 GAADEASRLLDQVKPTLPVLLANLVTVGQIGVTYNPSIEQLLVLLPPYLA
TH_4161|M.thermoresistible__bu GAADQAARLLAQVKPTVPVLLANLTTVGQIGVTYNPSLEQLLVLLPPYLA
Mflv_4533|M.gilvum_PYR-GCK GAADEASRLLTDVKPTLPVLLANLTTVGQVGVAYHPSLEQLLVLLPPYLA
MMAR_4705|M.marinum_M GAFNEASRLLDQIKPTLPVLLANLTTVGQIAVTYHASVEQILVLLPPFTA
MAV_0953|M.avium_104 ETANEASRLLDQIKPTLPVLLANLTTIGQIGITYHPSLEQLLVLLPSAVA
Mvan_4145|M.vanbaalenii_PYR-1 AFAQEVSALLNQVKPTLPILLANLSTVGQTLLTYNPAIEQLLVLFPGIIA
MAB_4148c|M.abscessus_ATCC_199 GASEQASQLFAGIRPSFPVLAASLANLGRVGVIYNKSIEQSLVILPALFS
Rv1971|M.tuberculosis_H37Rv PALGETRQLLERLQPTVPILLANLVSVGQVALTYHNDIEQLLVVFPMAIA
MUL_3923|M.ulcerans_Agy99 GAADEVRALFNRLRPTLPIVLANLVSVGEVAVAYHPSLEQLLVLFPQGTA
Mb0180|M.bovis_AF2122/97 PTADEVNAVFSGVRDSLPQTLANLEVVFDMLKRYHAGVEQLLVFLPQGAA
MLBr_02594|M.leprae_Br4923 PTADEVNTVFSDVRDSLPQTLANFEILIDMLKRYHHGVEQMLVFLPQGAS
:. :: :: :.* *.: : *: :** **.:* :
MSMEG_2860|M.smegmatis_MC2_155 ATQSYGSSLNNPTG---MALSEFSLTLGDPPACTVGFLPPSEWRSPADLT
TH_4161|M.thermoresistible__bu ATQSYGSSLNNPTG---MALSEFSLTLGDPPACTVGFLPPSEWRSPADLS
Mflv_4533|M.gilvum_PYR-GCK ATQSYGSSLNNPTG---MALSEFSLTLGDPPACSVGFLPPSSWRSPADLT
MMAR_4705|M.marinum_M AIQSF-LGTKSPVG---LATGAFNLIISDPPACTVGFLPPSQWRNPADTT
MAV_0953|M.avium_104 IEQAA-AAPNHPEG---TAQGDFALTIDDPPICTVGFMPPNTWRSPDDLS
Mvan_4145|M.vanbaalenii_PYR-1 AQQSFGLPQNSPTG---LPMGDFALTISDPNPCTVGFLPSTQWRSPEDET
MAB_4148c|M.abscessus_ATCC_199 ALLTVAGGAPMDEG---AKL-DFRLNLGDPPPCLTGFVPPPLMRTPADET
Rv1971|M.tuberculosis_H37Rv AEQAGILANLNTKQAYRGQYLSFNLNLNLPPPCTTGFLPAQQRRIPTFED
MUL_3923|M.ulcerans_Agy99 VTQAVGVAKRNTIQDYKGDYLVFNLNLNLPPPCTTGFLPAQQQRVPTFED
Mb0180|M.bovis_AF2122/97 IAQTVLTPTPGAAQ------LPLAPAINYPPPCLTGFLPASEWRSPADTS
MLBr_02594|M.leprae_Br4923 IAQTASATFPNMAA------LDLALSINQPPPCLTGFIPAAEWRSPADTS
: : :. * * .**:*. * *
MSMEG_2860|M.smegmatis_MC2_155 DVDTPDG-LYCKLPQDSPIG-VRGARNFPCMGQPGKRAPTVEICESDRPF
TH_4161|M.thermoresistible__bu DIDTPDG-LYCKLPQDSPIG-VRGARNFPCMGQPGKRAPTVEICESDRPF
Mflv_4533|M.gilvum_PYR-GCK DVDTPDG-LYCKLPQDSPIG-VRGTRNFPCMEQPGKRAPTVEICQSDRPF
MMAR_4705|M.marinum_M IVDTPDG-LYCKLPQDSPIG-VRGARNYPCMGQPGKRAPTVEICESDQPF
MAV_0953|M.avium_104 DIDTPDG-LYCKLPQDSPLS-VRGARNYPCMGHPGKRAPTVEICNSDQPF
Mvan_4145|M.vanbaalenii_PYR-1 TIDTPDG-LYCKLPQDSPMN-VRGARNYPCIEHPGKRAPTVELCNDPRGF
MAB_4148c|M.abscessus_ATCC_199 LRELPRD-MYCKTAQNDPSA-VRGARNYPCQEFPGKRAPTIQLCRDPRGY
Rv1971|M.tuberculosis_H37Rv YPDRPAGDLYCRVPQDSPFN-VRGARNIPCETVPGKRAPTVKLCESDAPY
MUL_3923|M.ulcerans_Agy99 YPNRAPGDLYCRVPQDAPFN-VRGARNLPCVTVPGKRAPTAKLCESNEVY
Mb0180|M.bovis_AF2122/97 PRPLPSG-TYCKIPQDAQLQ-VRGARNIPCVDVPGKRAATPKECRSKDPY
MLBr_02594|M.leprae_Br4923 LQPLPVG-TYCKIPMDTPANSVRGSRNIPCVDVPGKRAASPRECRDSKPY
. . **: . : ***:** ** *****.: . *.. :
MSMEG_2860|M.smegmatis_MC2_155 EPLAMRQHVTGPYPIDPNLIAQG--------VPPDDRVTLD-EHIYGPVE
TH_4161|M.thermoresistible__bu EPLAMRQHVTGPYPIDPNLIAQG--------VAPDSRVTSD-DQIFGPPA
Mflv_4533|M.gilvum_PYR-GCK EPLAMRQHVTGPYPIDPALIAQG--------VPPDDRVTLD-EQIYGPLE
MMAR_4705|M.marinum_M EPLAMRQHIFGTYPLDPNLIAQG--------IPPDDRANWNRERIFGPVE
MAV_0953|M.avium_104 MPLAMKQHVLGPYPLDPNLLAQG--------VPPDDRVTVN-DRIFGPVE
Mvan_4145|M.vanbaalenii_PYR-1 VPTAIRNHITGPYPFDPNLVSQG--------VPIDSTVEGP-DRIYLPPE
MAB_4148c|M.abscessus_ATCC_199 VPIGSNPWRGPPAPYDTPVEDPRN------ILPPNKFPNIP--PGAEPDP
Rv1971|M.tuberculosis_H37Rv LPLNDGYNWKG----DPNATVPG---------------------------
MUL_3923|M.ulcerans_Agy99 VPLNDGYNWKG----DPNSTLSGQPIPQLP--------------------
Mb0180|M.bovis_AF2122/97 VPLGTNPWFGDPNQILTCPAPGARCDQPVKPGLVIPAPSINTGLNPAPAD
MLBr_02594|M.leprae_Br4923 VPLGTNPWFGDPNQLLTCPAPGARCDQPVKPGLVIPAPTVDNGLNPAPAD
* .
MSMEG_2860|M.smegmatis_MC2_155 GTP-VDGD------------------------------------------
TH_4161|M.thermoresistible__bu GTP-PPADDTP---------------------------------------
Mflv_4533|M.gilvum_PYR-GCK GTP-MPAP------------------------------------------
MMAR_4705|M.marinum_M GTP-LPPGVGP------PEETVMPPGPLG---PPSPMG------------
MAV_0953|M.avium_104 GTP-LPPGAVPRGAPAGPRGENPPPGSVGAAVPPVPSSGPPALAPMSRMS
Mvan_4145|M.vanbaalenii_PYR-1 GTP-LPPGAVRAG-TPPVSEPGTPPFPVGAPAPMVPGAP-----------
MAB_4148c|M.abscessus_ATCC_199 GTPTLPPPSPP---------------------------------------
Rv1971|M.tuberculosis_H37Rv --------------------------------------------------
MUL_3923|M.ulcerans_Agy99 --------------------------------------------------
Mb0180|M.bovis_AF2122/97 QVQGTPPPVSDPLQRP----------------------------------
MLBr_02594|M.leprae_Br4923 RLPGTPPPVSDPLSQP----------------------------------
MSMEG_2860|M.smegmatis_MC2_155 --------------------------------------ALPVAPS-AHTA
TH_4161|M.thermoresistible__bu -----------------------------------PPGAAPASAS-AEAT
Mflv_4533|M.gilvum_PYR-GCK -----------------------------------APGAAPASVPGPVPA
MMAR_4705|M.marinum_M -EVSPIAPIDVPS-------TPPPAPSG-------APSAAPSGLG-RAAA
MAV_0953|M.avium_104 ADLPPIAPLDVPTPTELPPPPPPPAPAAPDQVDGAAPQAAPSAFA-GKAS
Mvan_4145|M.vanbaalenii_PYR-1 --PPAQPPILPPAPLLPGQGPLLPGQAVPVAPASGAVPVAPSAFG--SND
MAB_4148c|M.abscessus_ATCC_199 -----------------------------------APEVPPAGGDVMPVS
Rv1971|M.tuberculosis_H37Rv --------------------------------------------------
MUL_3923|M.ulcerans_Agy99 -------------------------------------PEPPLEQSAPPGP
Mb0180|M.bovis_AF2122/97 ----------------------------------GSGTVQCNGQQPNPCV
MLBr_02594|M.leprae_Br4923 ----------------------------------RSGSVQCNGQQPNTCV
MSMEG_2860|M.smegmatis_MC2_155 GDGPSVAIATYDPETGRYATPDGQVHHQADLATP-----GAQNWEDLLPR
TH_4161|M.thermoresistible__bu DGGPSVAFATYNPRTGQYATPDGLVHTRTDLVAP-----PAQNWTDLLPR
Mflv_4533|M.gilvum_PYR-GCK AGGPPVAVATYNPETGQVATPDGEVFTQSNLAAPNPAAGEGKSWQDMLPR
MMAR_4705|M.marinum_M RPGPSVAFAQYDPHTGSYVAPDGQVYRQTDLIIPD-TPKAPRTWKDMLPT
MAV_0953|M.avium_104 KPAPSVVVAKYDPRTGRYVGPDGKLYQQSDLVT----PKAPKTWKDMLPT
Mvan_4145|M.vanbaalenii_PYR-1 SGVPTVGTAQYNPQTGEYIAPDGSMNRVTNVASG----ANPKSWKDLLPM
MAB_4148c|M.abscessus_ATCC_199 AQSGPMAARAYDPKNGVFLDANNQPSVYAPALDRG---TEAETWADLMLG
Rv1971|M.tuberculosis_H37Rv ------------------LGS----------GQDIP-----QTWQTMLLP
MUL_3923|M.ulcerans_Agy99 APPAPIAAAEYDPATGTYMGPDGHLYTQANLGQTVPE---EQTWQTMLLP
Mb0180|M.bovis_AF2122/97 YTPTSGPSAVYSPASGELVGPDGVKYAVANSSTTG-----DDGWKEMLAP
MLBr_02594|M.leprae_Br4923 YTPSGPPSAVYSPQSGELVGPDGVAYAVENSTKTG-----DDGWKEMLAP
. * ::
MSMEG_2860|M.smegmatis_MC2_155 ----
TH_4161|M.thermoresistible__bu ----
Mflv_4533|M.gilvum_PYR-GCK ----
MMAR_4705|M.marinum_M ----
MAV_0953|M.avium_104 ----
Mvan_4145|M.vanbaalenii_PYR-1 ----
MAB_4148c|M.abscessus_ATCC_199 PKPV
Rv1971|M.tuberculosis_H37Rv PGS-
MUL_3923|M.ulcerans_Agy99 PKK-
Mb0180|M.bovis_AF2122/97 AS--
MLBr_02594|M.leprae_Br4923 AG--