For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAVWLITGASRGLGSAIARAALAEGHTVVAGARNVPAAAAELPHSDALTVIDLDVTNSAQVSAVADATAK RHGGIDVLVNNAGYSVLGAVEELTDTETRDMFEVNVFGLHRMTRAVLPHMRARGRGRILNIGSVGGFAAV ASSGLYGATKFAVEAITEALALELRGSGVSATVIEPGAFRTDFLSPRSVRFAATEIDAYADTAGAARTAF AALDGHQPGDPDKAAAAILAVAAADPAPLRVQLGRDCVARVERKLLDVCQELGRWRAIAESTDFSTSPAE STSGTRTVGAA
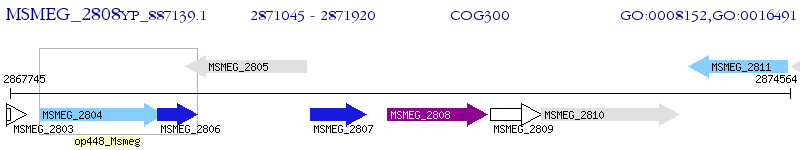
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2808 | - | - | 100% (291) | short-chain dehydrogenase/reductase SDR |
| M. smegmatis MC2 155 | MSMEG_0524 | - | 4e-59 | 47.37% (266) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2026 | - | 3e-57 | 44.60% (278) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2025 | fabG3 | 4e-30 | 41.76% (182) | 20-beta-hydroxysteroid dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0930 | - | 6e-51 | 41.45% (275) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2002 | fabG3 | 4e-30 | 41.76% (182) | 20-beta-hydroxysteroid dehydrogenase |
| M. leprae Br4923 | MLBr_00429 | - | 4e-20 | 33.33% (183) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_3405 | - | 4e-27 | 35.81% (229) | short chain dehydrogenase |
| M. marinum M | MMAR_1614 | - | 4e-51 | 42.39% (276) | short chain dehydrogenase |
| M. avium 104 | MAV_3827 | - | 6e-58 | 46.18% (275) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3670 | - | 6e-43 | 39.93% (273) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3423 | - | 6e-51 | 42.39% (276) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_5466 | - | 4e-51 | 41.61% (274) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2025|M.bovis_AF2122/97 --MSGRLIGKVALVSGGARG-MGASHVRAMVAEGAKVVFGDILDEEGKAV
Rv2002|M.tuberculosis_H37Rv --MSGRLIGKVALVSGGARG-MGASHVRAMVAEGAKVVFGDILDEEGKAV
MMAR_1614|M.marinum_M -------MST-WLITGCSTG-LGRALAEAVIDAGHHTVATARSVGGVADL
MUL_3423|M.ulcerans_Agy99 -------MST-WLITGCSTG-LGRALAEAVIDAGHHTVATARSVGGVADL
MAV_3827|M.avium_104 -------MAR-WLITGCSTG-LGREIAGAALQAGHRVVVTARRADAVRGF
MSMEG_2808|M.smegmatis_MC2_155 -------MAV-WLITGASRG-LGSAIARAALAEGHTVVAGARNVPAAAAE
Mflv_0930|M.gilvum_PYR-GCK -------MSKRWFVTGGTPGGFGVAFAEAALEAGDRVVITSRRPHELAAW
TH_3670|M.thermoresistible__bu -MGSNVWGSKVWFITGASRG-FGREWAIAALDRGDRVAATARDVSTLDDL
Mvan_5466|M.vanbaalenii_PYR-1 -MPYPTDNPATWFVTGASRG-LGAELVRQLLRRGDNVAATTRSAERLSAA
MAB_3405|M.abscessus_ATCC_1997 ---MDKITGKNIAITGAARG-IGYATATALLRRGARVVIGDRDVEALGSA
MLBr_00429|M.leprae_Br4923 MPIPAPSPEARAVVTGASQN-IGEALATELAAHGHSLIVIARREDVLVDL
::* : . :* . *
Mb2025|M.bovis_AF2122/97 AAELA--DAARYVHLDVTQPAQWTAAVDTAVTAFGGLHVLVNNAGILNIG
Rv2002|M.tuberculosis_H37Rv AAELA--DAARYVHLDVTQPAQWTAAVDTAVTAFGGLHVLVNNAGILNIG
MMAR_1614|M.marinum_M VQRSP--ERVLPLALDITEPDQITAAVQAAQQRFGGIDVLVNNAGYGYRA
MUL_3423|M.ulcerans_Agy99 AQRSP--ERVLPLALDITEPDQITAAMQAAQQRFGGIDVLVNNAGYGYRA
MAV_3827|M.avium_104 AEEFG--ELALPVALDVTDRDQIAAAVAAADAAFGGIDVLVNNAGHGYLS
MSMEG_2808|M.smegmatis_MC2_155 LPHS---DALTVIDLDVTNSAQVSAVADATAKRHGGIDVLVNNAGYSVLG
Mflv_0930|M.gilvum_PYR-GCK ADGYD--DRVLVVPLELTDAAQVQRVVQAAEEHFGGIDVLVNNAGRGWYG
TH_3670|M.thermoresistible__bu RDRFG--DAVLAMTLDVTDRAADFAAVQRAHDHFGRLDIVVNNAGYGHFG
Mvan_5466|M.vanbaalenii_PYR-1 LAGVDT-GRLLPLVVDLTDAAAVDGAVAAALDRFGRLDVVVNNAGYGFLG
MAB_3405|M.abscessus_ATCC_1997 VEGLKEEGNVDAHPLDVTDPASFKRFLEAASAG-GPIDVLVNNAGVMPIG
MLBr_00429|M.leprae_Br4923 AARLADKYRVAVEVRPADLTDPSERVKLADELTVRPISILCANAGTATFG
: :: *** .
Mb2025|M.bovis_AF2122/97 TIEDYALTEWQRILDVNLTGVFLGIRAVVKPMKEAGRGSIINISSIEGLA
Rv2002|M.tuberculosis_H37Rv TIEDYALTEWQRILDVNLTGVFLGIRAVVKPMKEAGRGSIINISSIEGLA
MMAR_1614|M.marinum_M AVEEGDDAEVRDLFETHFFGTVALIKAVLPDMRARRSGAIVNISSIAVAL
MUL_3423|M.ulcerans_Agy99 AVEEGDDAEVRDLFETHFFGTVALIKAVLPDMRARRSGAIVNISSIAVAL
MAV_3827|M.avium_104 AVEEGEDAEVRKLFDVNYFGAVDMIKAVLPGMRARGCGHIVNISSMTGLV
MSMEG_2808|M.smegmatis_MC2_155 AVEELTDTETRDMFEVNVFGLHRMTRAVLPHMRARGRGRILNIGSVGGFA
Mflv_0930|M.gilvum_PYR-GCK SIEGMDESSLRAMFELNFFAVLSVTRAVLPGMRARGSGWIVNVSSVAGLV
TH_3670|M.thermoresistible__bu FVEELTETEVRDQMETNFFGALWVTQAALPYLRRQRSGHIIQVSSIGGVS
Mvan_5466|M.vanbaalenii_PYR-1 AVEETTDAEIRQMLDVQVSGVWNVLRSAVPHLRAARSGHIINVSSILGLT
MAB_3405|M.abscessus_ATCC_1997 PFLSTSDRAIRSMLEVNLYGVINGCQLALPEMVARRRGQVVNIASVAGLM
MLBr_00429|M.leprae_Br4923 PVSALDPAGEKAQVQLNAVAVHDLTLAVLPGMIERKAGGILISGSAAGNS
. : .: : . .: : * :: .*
Mb2025|M.bovis_AF2122/97 GTVACHGYTATKFAVRGLTKSTALELGPSGIRVNSIHPGLVKT-------
Rv2002|M.tuberculosis_H37Rv GTVACHGYTATKFAVRGLTKSTALELGPSGIRVNSIHPGLVKT-------
MMAR_1614|M.marinum_M TPVGSGYYAAAKAAMEGMSGALHGELAPLGISVTVVEPGAFRTDFAG-RS
MUL_3423|M.ulcerans_Agy99 TPVGSGYYAAAKAAMEGMSGALHGELAPLGISVTVVEPGAFRTDFAG-RS
MAV_3827|M.avium_104 ANPPNAYYSSTKFALEAVTEALAAEVRPLGIKVTAIEPGAFRTDWAT-RS
MSMEG_2808|M.smegmatis_MC2_155 AVASSGLYGATKFAVEAITEALALELRGSGVSATVIEPGAFRTDFLSPRS
Mflv_0930|M.gilvum_PYR-GCK AAPGFGYYSATKFAIEAVTDALRDEVSTHGISVLAVEPGAFRTNAYAGFA
TH_3670|M.thermoresistible__bu AFADLGAYHASKWALEGLSQSLSLEVAPFGIHVTLIEPGGYATDWGGSS-
Mvan_5466|M.vanbaalenii_PYR-1 AVPGWGLYCAGKFALNGMSEALAAELAGFGIRVTIVEPGYFRTDFLTTES
MAB_3405|M.abscessus_ATCC_1997 AVPGMAVYNASKFGVVGLSRALSDEFAPQGVQVSAVLPTFTNTELIAGTD
MLBr_00429|M.leprae_Br4923 PIPYNATYAATKAFANTFSESLRGELHGSGVHVTLLAPGPVRTELPD---
* : * .: : *. *: . : * *
Mb2025|M.bovis_AF2122/97 ------------PMTDWVPEDIFQTALGRAAEPVEVSNLVVYLASD-ESS
Rv2002|M.tuberculosis_H37Rv ------------PMTDWVPEDIFQTALGRAAEPVEVSNLVVYLASD-ESS
MMAR_1614|M.marinum_M LVQSTRMINDYAATAGQRRKENDTMHGNQPGDPAKAAAAIITAVES-PAP
MUL_3423|M.ulcerans_Agy99 LVQSTRMINDYAATAGQRRKENDTMHGNQPGDPAKAAAAIITAVES-PAP
MAV_3827|M.avium_104 MKESGTPIADYTDVAA-RKDLIKQFADHLPGDPRKVAEAVLMVTGL-DEP
MSMEG_2808|M.smegmatis_MC2_155 VRFAATEIDAYADTAGAARTAFAALDGHQPGDPDKAAAAILAVAAA-DPA
Mflv_0930|M.gilvum_PYR-GCK NKPVAETIPSYHDMLEEVRAAFVEMDGVQPGDPRRGARAVIAAMAQ-DPP
TH_3670|M.thermoresistible__bu -ARRSATMPEYAESRAEADRVR-SQRVARPGDPKASAAALLKVVDA-EKP
Mvan_5466|M.vanbaalenii_PYR-1 LALPTAAGDAYPAIREMTEQHL-QLQGSQLGDPVKGAAAVIHVAVT-GGG
MAB_3405|M.abscessus_ATCC_1997 STGAQKPVEPEDIAAAVVRIIEKPRVQVTVPKPLGAVTTIVNSLPTGVRR
MLBr_00429|M.leprae_Br4923 --------------HDEASLVEKLVPDFLWISTEHTARLSLDALERNKMR
.. :
Mb2025|M.bovis_AF2122/97 ---YSTGAEFVVDGGTVAGLAHNDFGAVEVSSQPEWVT------------
Rv2002|M.tuberculosis_H37Rv ---YSTGAEFVVDGGTVAGLAHNDFGAVEVSSQPEWVT------------
MMAR_1614|M.marinum_M PAFLLLGPDALMAYRHIAQRRDVEITDWEQLTAGTNLTP-----------
MUL_3423|M.ulcerans_Agy99 PAFLLLGPDALMAYRHIAQRRDVEITDWEQLTAGTNLTP-----------
MAV_3827|M.avium_104 PLRLLLGRDVLKAMRDKIAAMSASIDEWEAVTKDVNFPGA----------
MSMEG_2808|M.smegmatis_MC2_155 PLRVQLGRDCVARVERKLLDVCQELGRWRAIAESTDFSTSPAESTSGTRT
Mflv_0930|M.gilvum_PYR-GCK PRRLVLGNSGYDAVIDALEQSLGDIRANETLSRSADFPT-----------
TH_3670|M.thermoresistible__bu PLRVFFGELPLQIVNADYQSRLQTWEEWQPIAVLAQG-------------
Mvan_5466|M.vanbaalenii_PYR-1 PLHQLLGSDAVSYAEAKIDALVADVKAGRDLALTTDHR------------
MAB_3405|M.abscessus_ATCC_1997 FMSKKTGTDRVFLDFDTSARTAYENRAGSSLGVTKPDGE-----------
MLBr_00429|M.leprae_Br4923 VVPGLTSKAMSVASRYAPRAIVASIVGNFYKKLGGG--------------
.
Mb2025|M.bovis_AF2122/97 ----
Rv2002|M.tuberculosis_H37Rv ----
MMAR_1614|M.marinum_M ----
MUL_3423|M.ulcerans_Agy99 ----
MAV_3827|M.avium_104 ----
MSMEG_2808|M.smegmatis_MC2_155 VGAA
Mflv_0930|M.gilvum_PYR-GCK ----
TH_3670|M.thermoresistible__bu ----
Mvan_5466|M.vanbaalenii_PYR-1 ----
MAB_3405|M.abscessus_ATCC_1997 ----
MLBr_00429|M.leprae_Br4923 ----