For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
HWFLPTYGDSRLIVGGGHGTPLGAAGGDREASIDYLASIVRAAERFGFTGALIPTGAWCEDAFVTAALLA RETTSLAFLVAFRPGLVSPTLSAQMAATFAKHAPGRILLNVVVGGEAHEQRAFGDHLDKEERYARCDEFL DVVRRLWAGETVTHRGKYIDVEEASLAAPPNPVPPLYFGGSSAAAGPVAARHSDVYLTWGEPPEAVREKI EWIRKLGEEQGRKLRFGIRLHTISRDTSEEAWAQADKLVAALDEETVRKAQEGLARSQSEGQRRMLALHE ANRLNGTWSDARSLEIAPNLWAGVGLVRGGAGTALVGSHTEVADRIAEYAEIGIDEFIFSGYPHLEELFW FGEGVVPILRKRGLFDAGALDCAPASIPFVGAAR*
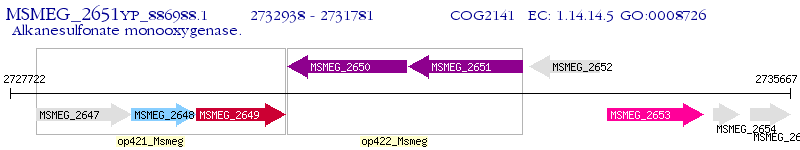
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2651 | - | - | 100% (385) | alkanesulfonate monooxygenase family protein |
| M. smegmatis MC2 155 | MSMEG_3850 | - | e-117 | 58.31% (367) | alkanesulfonate monooxygenase family protein |
| M. smegmatis MC2 155 | MSMEG_4539 | - | e-105 | 53.64% (371) | alkanesulfonate monooxygenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0137c | fgd2 | 7e-12 | 27.59% (174) | putative f420-dependant glucose-6-phosphate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_5136 | - | 0.0 | 81.15% (382) | alkanesulfonate monooxygenase |
| M. tuberculosis H37Rv | Rv0132c | fgd2 | 8e-12 | 27.59% (174) | putative f420-dependent glucose-6-phosphate dehydrogenase |
| M. leprae Br4923 | MLBr_00269 | - | 5e-07 | 27.78% (126) | putative F420-dependent glucose-6-phosphate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2219 | - | 1e-119 | 60.22% (367) | putative alkanesulfonate monooxygenase |
| M. marinum M | MMAR_4200 | fgd2 | 1e-126 | 60.93% (366) | F420-dependent glucose-6-phosphate dehydrogenase Fgd2 |
| M. avium 104 | MAV_4628 | - | 1e-131 | 61.75% (366) | alkanesulfonate monoxygenase family protein |
| M. thermoresistible (build 8) | TH_3352 | - | 0.0 | 83.94% (386) | PUTATIVE Alkanesulfonate monooxygenase |
| M. ulcerans Agy99 | MUL_4507 | fgd2 | 1e-125 | 60.66% (366) | F420-dependent glucose-6-phosphate dehydrogenase Fgd2 |
| M. vanbaalenii PYR-1 | Mvan_1200 | - | 0.0 | 84.16% (385) | alkanesulfonate monooxygenase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2651|M.smegmatis_MC2_155 --------------------------------MHWFLPTYGDSRL-----
TH_3352|M.thermoresistible__bu ----------------------------VTIKLHWFLPTYGDSRL-----
Mflv_5136|M.gilvum_PYR-GCK --------------------------MTHAIKLHWFLPTYGDSRH-----
Mvan_1200|M.vanbaalenii_PYR-1 MCTVAVLQIPLTQTADLPTPSGHHLVVTSAIKLHWFLPTYGDSRL-----
MMAR_4200|M.marinum_M ----------------------------MALSFHWFLPTYGDSRG-----
MUL_4507|M.ulcerans_Agy99 ----------------------------MALLFHWFLPTYGDSRG-----
MAV_4628|M.avium_104 ----------------------------MSLAFHWFLPTYGDSRN-----
MAB_2219|M.abscessus_ATCC_1997 ----------------------------MAAKFFWFLPTNGDSRS-----
Mb0137c|M.bovis_AF2122/97 -------------------------MTGISRRTFGLAAGFGAIGAGGLGG
Rv0132c|M.tuberculosis_H37Rv -------------------------MTGISRRTFGLAAGFGAIGAGGLGG
MLBr_00269|M.leprae_Br4923 --------------------------------------------------
MSMEG_2651|M.smegmatis_MC2_155 --------IVGGGHG-TPLGAAGGDREASIDYLASIVRAAERFGFTGALI
TH_3352|M.thermoresistible__bu --------IVGGGHG-TPAGAAGGDRDASIDYLASIVRAAETFGFTGALI
Mflv_5136|M.gilvum_PYR-GCK --------IVGGGHG-TPAGSAHSDRDASIDYLASIVRAAETFGFTGALI
Mvan_1200|M.vanbaalenii_PYR-1 --------IVGGGHG-TPAGAARGDRDASIDYLASIVRAAETFGFTGALI
MMAR_4200|M.marinum_M --------LVAGGHG-LPMS---GNRPATLRYLNQIAAAAESNGFEAVLT
MUL_4507|M.ulcerans_Agy99 --------LVAGGHG-LPMS---GNRPATLRYLNQIAAAAESNGFEAVLT
MAV_4628|M.avium_104 --------LVAGGHG-TPMS---GDRPATLRYLHQICSAAEDNGFEAVLT
MAB_2219|M.abscessus_ATCC_1997 --------IVGASHASTHHTVPEGYREPSLRYLGEVARAADRLGFEGVLT
Mb0137c|M.bovis_AF2122/97 GCSTRSGPTPTPEPASRGVGVVLSHEQFRTDRLVAHAQAAEQAGFRYVWA
Rv0132c|M.tuberculosis_H37Rv GCSTRSGPTPTPEPASRGVGVVLSHEQFRTDRLVAHAQAAEQAGFRYVWA
MLBr_00269|M.leprae_Br4923 -------------MAELRLGYKASAEQFAPRELVELGVAAEAHGMDSATV
. . . * **: *: .
MSMEG_2651|M.smegmatis_MC2_155 PTG--AWCED------AFVTAALLARETTSLAFLVAFRPGLVS--PTLSA
TH_3352|M.thermoresistible__bu PTG--AWCED------AFITAALLARETTSLGFLVAFRPGLVS--PTLSA
Mflv_5136|M.gilvum_PYR-GCK PTG--AWCED------AFITAALLARETSTLGFLVAFRPGLVS--PTLSA
Mvan_1200|M.vanbaalenii_PYR-1 PTG--AWCED------AFITAALLARETTSLAFLVAFRPGLVS--PTLSA
MMAR_4200|M.marinum_M PAG--LWCED------AWLTTAMLVGTTETLKFLVALRPGLVS--PTLAA
MUL_4507|M.ulcerans_Agy99 PAG--LWCED------AWLTTAMLVGTTKTLKFLVALRPGLVS--PTLAA
MAV_4628|M.avium_104 PTG--LWCED------AWLTTAMLVESTETLKFLVAFRPGLLS--PTLAA
MAB_2219|M.abscessus_ATCC_1997 PTG--TWCED------AWLTAAGLLDQTERLKFLVAFRPGLVP--PTLAA
Mb0137c|M.bovis_AF2122/97 SDHLQPWQDNEGHSMFPWLTLALVGNSTSSILFGTGVTCPIYRYHPATVA
Rv0132c|M.tuberculosis_H37Rv SDHLQPWQDNEGHSMFPWLTLALVGNSTSSILFGTGVTCPIYRYHPATVA
MLBr_00269|M.leprae_Br4923 SDHFQPWRHQGGHASFSLSWMTAVGERTNRILLGTSVLTPTFRYNPAVIG
. * .: . : : * : : ... *: .
MSMEG_2651|M.smegmatis_MC2_155 QMAATFAKHAPGRILLNVVVGGEAHEQRA--FGDHLDKEERYARCDEFLD
TH_3352|M.thermoresistible__bu QMAATFARHAPGRILLNVVVGGESHEQRA--FGDHLDKDERYRRADEFLE
Mflv_5136|M.gilvum_PYR-GCK QMAATFARHAPGRLLLNVVVGGEAHEQRS--FGDHLDKDGRYARADEFLD
Mvan_1200|M.vanbaalenii_PYR-1 QMAATFARHAPGRILLNVVVGGEAHEQRS--FGDYLDKEGRYARADEFLD
MMAR_4200|M.marinum_M QMATTFQQQSGGRLLLNVVTGGEAHEQRA--YGDFLDKQQRYRRTGEFLH
MUL_4507|M.ulcerans_Agy99 QMATTFQQQSGGRLLVNVVTGGEAHEQRA--YGDFLDKQQRYRRTGEFLH
MAV_4628|M.avium_104 QMAGTFQRHSQGRLLLNVVTGGEPHEQRA--YGDFLDKEARYARTAEFLH
MAB_2219|M.abscessus_ATCC_1997 QQAATLQRFSNGRLLLNVVSGGDDAEQRR--FGDWLTHDERYERTGEFLQ
Mb0137c|M.bovis_AF2122/97 QAFASLAILNPGRVFLGLGTGKRLNEQAA--TDTFGNYRERHDRLIEAIV
Rv0132c|M.tuberculosis_H37Rv QAFASLAILNPGRVFLGLGTGERLNEQAA--TDTFGNYRERHDRLIEAIV
MLBr_00269|M.leprae_Br4923 QAFATMGCLYPNRVFLGVGTGEALNEVATGYQGAWPEFKERFARLRESVR
* :: .*:::.: * * . *. * * :
MSMEG_2651|M.smegmatis_MC2_155 VVRRLWAG-ETVTHRGKYIDVEEASLAAPPNPVPPLYFGGSSAAAGPVAA
TH_3352|M.thermoresistible__bu VVRRLWAG-ETVTLHGRHIRVEDAALPTLPDPVPPLYFGGSSTAAGPVAA
Mflv_5136|M.gilvum_PYR-GCK VVRRLWDG-QTVSLDGEHIRVENASLPTVPAPVPPLYFGGSSAAAGPVAA
Mvan_1200|M.vanbaalenii_PYR-1 VVRRLWDG-QTVTLDGEHIRVEGASLATLPDPAPPLYFGGSSAAAGPVAA
MMAR_4200|M.marinum_M VVRQLWASGQPVTFEGEHIHVHQAQLGRLPDPPPPVFFGGSSESAGAVAA
MUL_4507|M.ulcerans_Agy99 VVRQLWASGQPVTFEGEHIHVHQAQLGRLPDPPPPVFFGGSSESAGAVAA
MAV_4628|M.avium_104 VVRQLWTSPQPVTFAGEHIRVEGAQLNNPPDPIPAVFFGGSSASAGPVAA
MAB_2219|M.abscessus_ATCC_1997 IVRDIWRG-SPVDFSGKHYTVTDARVSEPPSPLPELYFGGSSPAALPIAA
Mb0137c|M.bovis_AF2122/97 LIRQLWSG-ERISFTGHYFRTDELKLYDTPAMPPPIFVAASGPQSATLAG
Rv0132c|M.tuberculosis_H37Rv LIRQLWSG-ERISFTGHYFRTDELKLYDTPAMPPPIFVAASGPQSATLAG
MLBr_00269|M.leprae_Br4923 LMRELWRG-DRVDFDGDYYQLKGASIYDVPEGGVPIYIAAGGPEVAKYAG
::* :* . . : * : : * ::..... *.
MSMEG_2651|M.smegmatis_MC2_155 RHSDVYLTWGEPPEAVREKIEWIRKLGEEQGRKLRFGIRLHTISRDTSEE
TH_3352|M.thermoresistible__bu RHADVYLTWGEPPDAVREKIDWIRTLAAAEGRRVRFGIRLHTISRDTADE
Mflv_5136|M.gilvum_PYR-GCK RHADVYLTWGEPPAAVAEKVEWIRRAAAGQGRQLRFGIRLHVITRDTAGA
Mvan_1200|M.vanbaalenii_PYR-1 RHADVYLTWGEPPAAVADKVDWIRRLAAKQGRQVRFGIRLHVITRDAADE
MMAR_4200|M.marinum_M RYADTYLTWGEPLSAVGKKIDWIRGLAADHGRALSYGLRIHVITRDTAEE
MUL_4507|M.ulcerans_Agy99 RYADTYLTWGEPLSAVGKKIDWIRGLAADHGRALSYGLRIHVITRDTAEE
MAV_4628|M.avium_104 KHSDVYLTWGEPLTAVAKKLDWVRGLAVDAGRILQYGLRIHVISRDTSAA
MAB_2219|M.abscessus_ATCC_1997 DHVDVYLTWGEPPAAAAAKIEAVRALAKERGRTLRFGIRLHTISRDTSEA
Mb0137c|M.bovis_AF2122/97 RYGDGWIAQAR-------DINDAKLLAAFAAGAQAAGRDPTTLGKRAELF
Rv0132c|M.tuberculosis_H37Rv RYGDGWIAQAR-------DINDAKLLAAFAAGAQAAGRDPTTLGKRAELF
MLBr_00269|M.leprae_Br4923 RAGEGFVCTSGK----GEELYTEKLIPAVLEGAAVAGRDADDIDKMIEIK
: :: . .: : * : :
MSMEG_2651|M.smegmatis_MC2_155 AWAQADKLVAALDEETVRKAQEGLARSQSEGQRRMLALHEANRLNGTWSD
TH_3352|M.thermoresistible__bu AWAQADKLIGALDEHTVRAAQAGLRRSESEGQRRMLALHQANRADGSWRS
Mflv_5136|M.gilvum_PYR-GCK AWAEADRLVGALDDETVRNAQEGLGRSQSEGQRRMSALHQAHRANGSWHD
Mvan_1200|M.vanbaalenii_PYR-1 AWSQADKLVSALDDETVRSAQAGLGRSQSEGQRRMLALHEAHRADGSWHD
MMAR_4200|M.marinum_M AWRVADRLLAGIDPADIERMQANLARSESEGQRRMAELHGG--------A
MUL_4507|M.ulcerans_Agy99 AWRVADRLLAGIDPADIERMQANLARSESEGQRRMAELHGG--------V
MAV_4628|M.avium_104 AWAEADRLLSAIDPADIERVQASLARSESEGQRRMRELHGG--------D
MAB_2219|M.abscessus_ATCC_1997 AWAVANSLVGELTPDQIAKATALHSKSESEGQRRMTALHGG--------R
Mb0137c|M.bovis_AF2122/97 AVVGDD-KAAARAADLWRFTAGAVDQPN----PVEIQRAAE---------
Rv0132c|M.tuberculosis_H37Rv AVVGDD-KAAARAADLWRFTAGAVDQPN----PVEIQRAAE---------
MLBr_00269|M.leprae_Br4923 MSYDPDPEQALSNIRFWAPLSLAAEQKHSIDDPIEMEKVAD---------
: . : .
MSMEG_2651|M.smegmatis_MC2_155 ARSLEIAPNLWAGVGLVRGGAGTALVGSHTEVADRIAEYAEIGIDEFIFS
TH_3352|M.thermoresistible__bu ARSLEVAPNLWAGVGLVRGGAGTALVGSHAEVADRIAEYAAAGIEEFIFS
Mflv_5136|M.gilvum_PYR-GCK ARTLEVAPNLWTGVGLVRGGAGTALVGSHSEVADRIAEYAEVGIDEFIFS
Mvan_1200|M.vanbaalenii_PYR-1 ARSLEIAPNLWAGVGLVRGGAGTALVGSHTEVADRIAEYAEAGIDEFVFS
MMAR_4200|M.marinum_M LDQLEIAPNLWAGVGLVRGGAGTALVGSHQEVAERIIEYARLGISHFILS
MUL_4507|M.ulcerans_Agy99 LDQLEIAPNLWAGVGLVRGGAGTALVGSHQEVAERIIEYARLGISHFILS
MAV_4628|M.avium_104 SGKLLVGPNLWAGVGLVRGGAGTALVGSHAEVAERLAEYAKLGITHFILS
MAB_2219|M.abscessus_ATCC_1997 LDKLEIYPNLWAGVGLVRGGAGTALVGSHEEVANLISEYHALGFDEFILS
Mb0137c|M.bovis_AF2122/97 SNPIEKVLANWAVG---------TDPGVHIGAVQAVLDAGA--VPFLHFP
Rv0132c|M.tuberculosis_H37Rv SNPIEKVLANWAVG---------TDPGVHIGAVQAVLDAGA--VPFLHFP
MLBr_00269|M.leprae_Br4923 ALPIEQVAKRWIVV---------SDPDEAVARVGQYVTWGLN-HLVFHAP
: * : . . : .
MSMEG_2651|M.smegmatis_MC2_155 GYPHLEELFWFGEGVVPILRKRGLFD-AGALDCAPASIPFVGAAR----
TH_3352|M.thermoresistible__bu GYPHLEELFWFGEGVVPILRSRNLFDGAGDRSPVPAAIPFVGAAR----
Mflv_5136|M.gilvum_PYR-GCK GYPHLEELYWLGEGVVPLLRERGLFE-AGPKQAASASIPFVGGTGRGRI
Mvan_1200|M.vanbaalenii_PYR-1 GYPHLEELFWFGEGVVPILRDRGLFT-ADVKGAAASSIPFVGAAR----
MMAR_4200|M.marinum_M GYPHLEEAYWFGEGVLPILERKRLWHRPHGSSPHSAALPVAHSSVN---
MUL_4507|M.ulcerans_Agy99 GYPHLEEAYWFGEGVLPILERKRLWHRPHGSSPHSAALPVAHSSVN---
MAV_4628|M.avium_104 GYPHLEEAYWFGEGVLPLLERMGLWRHPNRRSRPAATTPFAVASAH---
MAB_2219|M.abscessus_ATCC_1997 GYPHLEEAYWFAEGVLPLLKKRGALAA----------------------
Mb0137c|M.bovis_AF2122/97 QDDPITAIDFYRTNVLPELR-----------------------------
Rv0132c|M.tuberculosis_H37Rv QDDPITAIDFYRTNVLPELR-----------------------------
MLBr_00269|M.leprae_Br4923 GHNQRRFLELFEKDLAPRLRRLG--------------------------
.: * *.