For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VESAFRSADIANAPTACSAVQVNRPRLPASPADQIRHLSMTTRTLRIAMNGVTGRMGYRQHLLRSILPLR ESGLTADDGSRVAVEPILVGRSADKLADMAAEHGIEHWTTDVASVIADPSVEVYFDAQVTSRRFEALTAA IKAGKHVYTEKPTAETLTEAVELARMAQNAGITAGVVHDKLYLPGLVKLRRLVDEGFFGRILSMRGEFGY WVFEGDGQPAQRPSWNYRAQDGGGITVDMFCHWNYVMEGILGKVETVTAKAVTHIPTRWDEAHQPYPVTA DDAVYGIFDISGGIVAQINSSWAVRVFRDELVEFQVDGTHGSAVAGLRRCRAQHRVHTPKPVWNPDLPAT EQFRDQWLEVPDNAVLDNGFKLQWEEYLRDVLAARPHRYDLLSAARGVQLAELGLKSSTEGRRLDVPEIV L
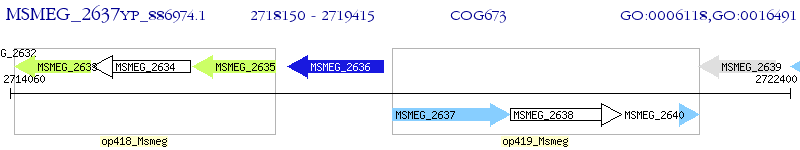
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2637 | - | - | 100% (421) | oxidoreductase, Gfo/Idh/MocA family protein |
| M. smegmatis MC2 155 | MSMEG_6717 | - | 4e-08 | 25.13% (199) | oxidoreductase, Gfo/Idh/MocA family protein |
| M. smegmatis MC2 155 | MSMEG_3300 | - | 4e-08 | 25.95% (185) | oxidoreductase, Gfo/Idh/MocA family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2611 | - | 2e-08 | 25.62% (160) | oxidoreductase domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2306c | - | 3e-09 | 23.84% (172) | putative oxidoreductase |
| M. marinum M | MMAR_2628 | - | 3e-07 | 24.60% (187) | hypothetical protein MMAR_2628 |
| M. avium 104 | MAV_2643 | - | 4e-15 | 28.64% (220) | oxidoreductase family protein, NAD-binding Rossmann fold |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3970 | - | 7e-10 | 26.95% (167) | oxidoreductase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2611|M.gilvum_PYR-GCK -------------------------------------MT-----LRIGVL
Mvan_3970|M.vanbaalenii_PYR-1 -------------------------------------MS-----LRIGIL
MMAR_2628|M.marinum_M -------------------------------------MTGHGSPLRIGIL
MAB_2306c|M.abscessus_ATCC_199 -------------------------------------MP---ESIRWGIV
MAV_2643|M.avium_104 -------------------------------------MT-----LRVGII
MSMEG_2637|M.smegmatis_MC2_155 MESAFRSADIANAPTACSAVQVNRPRLPASPADQIRHLSMTTRTLRIAMN
:. :* .:
Mflv_2611|M.gilvum_PYR-GCK GASRIAEA-------------AIVEPAR-ELGFRLVAVAARDPQRANIFA
Mvan_3970|M.vanbaalenii_PYR-1 GASRIAAS-------------AIVEPAR-ELGHRLIAVAARDPLRTQAFA
MMAR_2628|M.marinum_M GAARIAPQ-------------ALVKPAQGNVEAVVAAVAARDGARAQAFA
MAB_2306c|M.abscessus_ATCC_199 GPGRIAG--------------NVARDFPLTPRAELAAVASRSLPRAQDFA
MAV_2643|M.avium_104 GVG-WGAR-------------VQVPGFRAAKGFEPVALCARTPDRLERAA
MSMEG_2637|M.smegmatis_MC2_155 GVTGRMGYRQHLLRSILPLRESGLTADDGSRVAVEPILVGRSADKLADMA
* : .* : *
Mflv_2611|M.gilvum_PYR-GCK DTYGVERVVPTYQDVVTDPEVDVVYNPLANSLHAPWNLAAVAAGKPVLSE
Mvan_3970|M.vanbaalenii_PYR-1 DKYGVERVLASYQDVVTDPQVDIVYNPLANALHAPWNLAAVAAGKPVLTE
MMAR_2628|M.marinum_M AKHGISRVHSSYEALIADPTLDAVYIPLPNGLHGRWTQAALTAGKHVLCE
MAB_2306c|M.abscessus_ATCC_199 GAHQIPRAYGSYRELLADPDIDAVYIATPHPQHRQTAIAALTAGKAILVE
MAV_2643|M.avium_104 AKLGIEQTSTDWQSFVTRDDLDVISVATPTVLHHEMTIAALAAGKPVLCE
MSMEG_2637|M.smegmatis_MC2_155 AEHGIEHWTTDVASVIADPSVEVYFDAQVTSRRFEALTAAIKAGKHVYTE
: : .:: :: . : **: *** : *
Mflv_2611|M.gilvum_PYR-GCK KPFARNRAEAARVASAADAAGVTVMEGFHYLFHPVTQRALALAGDGTLG-
Mvan_3970|M.vanbaalenii_PYR-1 KPYARDRGEAARVAAAADAAGVTVMEGFHYLFHPVTQRALSLAGDGTLG-
MMAR_2628|M.marinum_M KPFTANAAEADEIAQLAATTDRVVMEAFHYRYHPLTLRIEQILASGELG-
MAB_2306c|M.abscessus_ATCC_199 KTFTATVSGAQEVIDIARARGVFAMEAMWTRFQPAIVAAKDLVDDGAIG-
MAV_2643|M.avium_104 KPLAGDLEAARHMVRAATDSKLPTACCFENRWNPDWLAVADKVRSGFLGS
MSMEG_2637|M.smegmatis_MC2_155 KPTAETLTEAVELARMAQNAGITAGVVHDKLYLPGLVKLRRLVDEGFFG-
*. : * .: * . : * .* :*
Mflv_2611|M.gilvum_PYR-GCK -ELRRVEVRMGMPEPK-------PDDPRWSLELAGGALMDLGCYGVHVLR
Mvan_3970|M.vanbaalenii_PYR-1 -ELTRIEVRMGMPAPQ-------SDDPRWSLELAGGALMDLGCYGLHVMR
MMAR_2628|M.marinum_M -TLQRVETSMCFPLPK-------FSDIRYNYALAGGATMDAGCYAVHMAR
MAB_2306c|M.abscessus_ATCC_199 -EVRQVQADLGVDRPFD------REDRLFNPALGGGALLDLGVYVVSLAQ
MAV_2643|M.avium_104 QYLARVSRSASYWHPSHP----LQAHWMYDREQGGGYLAGMLVHDLDFLT
MSMEG_2637|M.smegmatis_MC2_155 -RILSMRGEFGYWVFEGDGQPAQRPSWNYRAQDGGGITVDMFCHWNYVME
: : : .** . : .
Mflv_2611|M.gilvum_PYR-GCK RFG-------------VPSILSATAVQRSAGVDATLDAEVVFPSGLTGLT
Mvan_3970|M.vanbaalenii_PYR-1 RFG-------------TPSVLAATAVQRTPGVDERFDAELVFPSGLTGLT
MMAR_2628|M.marinum_M TFGGG-----------TPEVVSAQAKRRDSEVDRAMTAELRFPAGHSGRI
MAB_2306c|M.abscessus_ATCC_199 YFLG------------DPAAVVAHGSLFPTGVDAEAGLLLRYDDGRTATL
MAV_2643|M.avium_104 SLLGRPVSVCAEVRTSDPVRELPDGGTLPVTADDTAALLMRMESGVTVIL
MSMEG_2637|M.smegmatis_MC2_155 GILGKVETVTAKAVTHIPTRWDEAHQPYPVTADDAVYGIFDISGGIVAQI
: * .* . *
Mflv_2611|M.gilvum_PYR-GCK ANSMVQ--DDFS-FTLRLIGSRGEAFVHNFIKPRDDDR----------LT
Mvan_3970|M.vanbaalenii_PYR-1 TNSMVD--DEYS-FTLRLIGTRGEAFVHDFIKPHDDDR----------LT
MMAR_2628|M.marinum_M QCSMWS--SHLLNMSARVVGDRGELRVLNPVVPQLFHR----------LT
MAB_2306c|M.abscessus_ATCC_199 LCSLLH--HTPG--QARIFGTEGWIDILPRFHHPREIV----------LH
MAV_2643|M.avium_104 SLSVMG--AHADHYRLELFGSDGTIIGDGDLRSAAYSLGAATDAGLSPLT
MSMEG_2637|M.smegmatis_MC2_155 NSSWAVRVFRDELVEFQVDGTHGSAVAGLRRCRAQHRVHTPKPVWNPDLP
* .: * * *
Mflv_2611|M.gilvum_PYR-GCK VQTDDGTRVERFG------TRASYTYQLEFFARHVLHG--APLPLDTTDA
Mvan_3970|M.vanbaalenii_PYR-1 LHTEDGTTVERHG------TRTSYTYQLEAFTDHVERG--TPIPLDTADA
MMAR_2628|M.marinum_M VQSAIGRRVERFP------RRASYAYQLDAFTGAILRG--EAVKTTPQDA
MAB_2306c|M.abscessus_ATCC_199 RKGAAPEPMVRPP------VGGGYSHELAEVTECLLNGRAQSSVMPLADT
MAV_2643|M.avium_104 VDEREPAHPERLPGGLAGHASRAMALMLQDWLPAFDGG--PSGAATFEDG
MSMEG_2637|M.smegmatis_MC2_155 ATEQFRDQWLEVPDN--AVLDNGFKLQWEEYLRDVLAAR--PHRYDLLSA
. . . . . .
Mflv_2611|M.gilvum_PYR-GCK VENMAVIDEIYRAAGLPVR--------
Mvan_3970|M.vanbaalenii_PYR-1 VANMALIDEAYRAAGMSPR--------
MMAR_2628|M.marinum_M VQNMTVIDAIYRAAGLPLRNPS-----
MAB_2306c|M.abscessus_ATCC_199 LAVQRVLNTACEQLGVDHTEDPADLD-
MAV_2643|M.avium_104 LLSLAVIDAAHRSAEGGGWEQVQV---
MSMEG_2637|M.smegmatis_MC2_155 ARGVQLAELGLKSSTEGRRLDVPEIVL
: : .