For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AAVTLQDVAVRAGVSQATASRVLNGSARIPGEGVADRVRAAARELGYVPNAQAQALARSSTGLLGLIVHD IADPYFSSIVRGVQTAARTARKQVLLASTDRDFDIEREAVSTFIAHRADAIVLAGSRQSGDLDRDIETEF GRYRDNGGRVVVIGQPLAFGGAVEPENHLGAAQLAEALVKSGHTQFAVIGGPANIRTAVDRRNGFVEALG RRGLTPLVEVSGDFTRDGGYSAARRLAGALQLSPGARGEPVCVFAVTDVMAIGAIAAWRELGLSVPDDVG IAGFDDIPTLRDHTPALTTVVLPLQDIGVRAVELALRTDADCDDLRERIPGRVVLRDSTRLP*
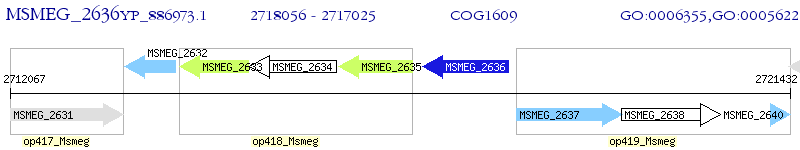
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2636 | - | - | 100% (343) | regulatory protein, LacI |
| M. smegmatis MC2 155 | MSMEG_1708 | - | 3e-36 | 34.21% (342) | ribose operon repressor, putative |
| M. smegmatis MC2 155 | MSMEG_2989 | - | 9e-34 | 30.41% (342) | LacI family protein transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3606c | - | 2e-11 | 24.50% (347) | transcriptional regulatory protein LacI-family |
| M. gilvum PYR-GCK | Mflv_1457 | - | 5e-15 | 25.29% (344) | periplasmic binding protein/LacI transcriptional regulator |
| M. tuberculosis H37Rv | Rv3575c | - | 2e-11 | 24.50% (347) | transcriptional regulatory protein LacI-family |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2023c | - | 1e-05 | 39.29% (56) | LacI family transcriptional regulator |
| M. marinum M | MMAR_5070 | - | 2e-11 | 24.41% (340) | PurR family transcriptional regulator |
| M. avium 104 | MAV_0585 | - | 1e-10 | 24.20% (343) | transcriptional regulator, LacI family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4146 | - | 2e-11 | 24.41% (340) | PurR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_2545 | - | 2e-22 | 28.21% (358) | periplasmic binding protein/LacI transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5070|M.marinum_M --MSPTPRRRATLASLAAELKVSRSTISNAFNRPDQLS-AELRERVLATA
MUL_4146|M.ulcerans_Agy99 --MSPTPRRRATLASLAAELKVSRSTISNAFNRPDQLS-VELRERVLATA
MAV_0585|M.avium_104 --MSPTPRRRATLASLADELKVSRTTVSNAFNRPDQLS-ADLRERVLATA
Mb3606c|M.bovis_AF2122/97 --MSPTPRRRATLASLAAELKVSRTTVSNAFNRPDQLS-ADLRERVLATA
Rv3575c|M.tuberculosis_H37Rv --MSPTPRRRATLASLAAELKVSRTTVSNAFNRPDQLS-ADLRERVLATA
Mflv_1457|M.gilvum_PYR-GCK MSRSPTPRRRATLASLAAELKVSRTTVSNAYNRPDQLS-AELRERVLSTA
Mvan_2545|M.vanbaalenii_PYR-1 ----MIPKQTVNMTDVARRSGVSIASVSRALRGESGVS-AATRDRILSAA
MSMEG_2636|M.smegmatis_MC2_155 -------MAAVTLQDVAVRAGVSQATASRVLNGSARIPGEGVADRVRAAA
MAB_2023c|M.abscessus_ATCC_199 -------------------MGLSKTTVADALGGSGRVS-EATREKTRLVA
:* :: : . :. :: .*
MMAR_5070|M.marinum_M KRMGYPGPDPVARSLRTRKAGAVGLVMAEPLTYFFSDPAARDFVTGVAQS
MUL_4146|M.ulcerans_Agy99 KRMGYPGPDPVARSLRTRKAGAVGLVMAEPLTYFFSDPAARDFVTGVAQS
MAV_0585|M.avium_104 KRLGYPGPDPVARSLRTRKAGAVGLVMAEPLTYFFSDPAARDFVTGVAQS
Mb3606c|M.bovis_AF2122/97 KRLGYAGPDPVARSLRTRKAGAVGLVMAEPLTYFFSDPAARDFVAGVAQS
Rv3575c|M.tuberculosis_H37Rv KRLGYAGPDPVARSLRTRKAGAVGLVMAEPLTYFFSDPAARDFVAGVAQS
Mflv_1457|M.gilvum_PYR-GCK KRLGYPGPDPVARSLRTRRAGAVGLVITEPLNYSFSDPAALDFVAGLAES
Mvan_2545|M.vanbaalenii_PYR-1 RELAYVVS-PEASRLSGGPTGRVGVIVPRVDAWFYST-----ILAGIADE
MSMEG_2636|M.smegmatis_MC2_155 RELGYVPN-AQAQALARSSTGLLGLIVHDIADPYFSS-----IVRGVQTA
MAB_2023c|M.abscessus_ATCC_199 ERMGYVSN-RAARLLRRNETGALGLYIPGDVRNLSFY---MELAFGAADE
..:.* * * :* :*: : : *
MMAR_5070|M.marinum_M CEELGQGLLLVAVGASRS-LEDGTAAVLNAGVDGFVVYSVRDD-------
MUL_4146|M.ulcerans_Agy99 CEELGQGLLLVAVGASRS-LEDGTAAVLNAGVDGFVVYSVRDD-------
MAV_0585|M.avium_104 CEALGQGLLLVAVGPSRS-LDEGTAAVLSAGVDGFVVYSVCDD-------
Mb3606c|M.bovis_AF2122/97 CEELGQGLQLVSVGSSRS-LADGTAAVLGAGVDGFVVYSVGDD-------
Rv3575c|M.tuberculosis_H37Rv CEELGQGLQLVSVGSSRS-LADGTAAVLGAGVDGFVVYSVGDD-------
Mflv_1457|M.gilvum_PYR-GCK CEAVGQGLLLVAVGPNRS-VAEGSAAVLAAGVDGFVVYSASDD-------
Mvan_2545|M.vanbaalenii_PYR-1 FDTVGMDLILCTLPTSAARHRFFEALPLRRKVDAAVVVSLPLS-------
MSMEG_2636|M.smegmatis_MC2_155 ARTARKQVLLASTDRDFDIEREAVSTFIAHRADAIVLAGSRQSGDLDRDI
MAB_2023c|M.abscessus_ATCC_199 SAALGLDLTLIGRHLTGE------NRSRPPQVDGIIAVDPVPG-------
: * .*. : . .
MMAR_5070|M.marinum_M DPYLQVVLQRRLPVVVVDQPKELAG---VSRVGIDDRAAMRKIADYVLGL
MUL_4146|M.ulcerans_Agy99 DPYLQVVLQRRLPVVVVDQPKELAG---VSRVGIDDRAAMRKIADYVLSL
MAV_0585|M.avium_104 DPYLQAVLQRRLPVVVVDQPKGLTG---VSRVGIDDRAAMRELADYVLGL
Mb3606c|M.bovis_AF2122/97 DPYLQVVLQRRLPVVVVDQPKDLSG---VSRVGIDDRAAMRELAGYVLGL
Rv3575c|M.tuberculosis_H37Rv DPYLQVVLQRRLPVVVVDQPKDLSG---VSRVGIDDRAAMRELAGYVLGL
Mflv_1457|M.gilvum_PYR-GCK DPYLPAVMERHLPVVVVDQPRDVPG---ASQVRIDDRAAMRSLAEHVIDL
Mvan_2545|M.vanbaalenii_PYR-1 ARERTRLDQLAVPTVFVGGHRTEDH---RPWVGIDDERAAQQAVGHLLRI
MSMEG_2636|M.smegmatis_MC2_155 ETEFGRYRDNGGRVVVIGQPLAFGG---AVEP--ENHLGAAQLAEALVKS
MAB_2023c|M.abscessus_ATCC_199 DAALESLLTSNVPIVTVGRILDKDVSHHAAEIEIDHYRMVTLLLDAIRAA
* :. :. :
MMAR_5070|M.marinum_M GHRQIGLLTMRLGRDRRQD----LVDAERLKSPTFDVQRERITGVWEAMT
MUL_4146|M.ulcerans_Agy99 GHRQIGLLTMRLGRDRRQD----LVDAERLKSPTFDVQRERITGVWEAMT
MAV_0585|M.avium_104 GHREIGLLTMRLGRDRRQG----LVDAERLRSPAFDVQRERITGVWEAMT
Mb3606c|M.bovis_AF2122/97 GHRELGLLTMRLGRDRRQD----LVDAERLRSPTFDVQRERIVGVWEAMT
Rv3575c|M.tuberculosis_H37Rv GHRELGLLTMRLGRDRRQD----LVDAERLRSPTFDVQRERIVGVWEAMT
Mflv_1457|M.gilvum_PYR-GCK GHREIGLLTMRLGRDWPHGPGPALADPERVRAPNFHVQCERIHGVYDAMT
Mvan_2545|M.vanbaalenii_PYR-1 GHHDIAMIQAADDTDIPWA-----------------TDQARIRGFHQTMQ
MSMEG_2636|M.smegmatis_MC2_155 GHTQFAVIGGPANIRTAVD---------------------RRNGFVEALG
MAB_2023c|M.abscessus_ATCC_199 GGKRPALAALDQGADSSYV-------------------FDVSRGYQEWCA
* .: . * :
MMAR_5070|M.marinum_M AAGVDPESLTVVESYEHLPTSGGDAAKVALEANP----RITALMCTADIL
MUL_4146|M.ulcerans_Agy99 AAGVDPESLTVVESYEHLPTSGGDAAKVALEANP----RITALMCTADIL
MAV_0585|M.avium_104 AAGVDPDSLTVVESYEHLPESGGAAAKVALEANP----RITALMCTADIL
Mb3606c|M.bovis_AF2122/97 AAGVDPDSLTVVESYEHLPTSGGTAAKVALQANP----RLTALMCTADIL
Rv3575c|M.tuberculosis_H37Rv AAGVDPDSLTVVESYEHLPTSGGTAAKVALQANP----RLTALMCTADIL
Mflv_1457|M.gilvum_PYR-GCK AAGLDPASLTLVESYEHLPTSGGAGADIALEANP----RITALMCTADVL
Mvan_2545|M.vanbaalenii_PYR-1 EAGLADPTVVTVK---WSVDGGSHGMETLLSRSR----PPTAVFCHSDEI
MSMEG_2636|M.smegmatis_MC2_155 RRGLTPLVEVSGDFTRDGGYSAARRLAGALQLSPGARGEPVCVFAVTDVM
MAB_2023c|M.abscessus_ATCC_199 KAGVVP--LATRLSATPTDVELAQAIRGIVEHQG-----IDALVVGAQGL
*: . . :. . .:. :: :
MMAR_5070|M.marinum_M ALSAMDYLRGHGIYVPGQMTVTGFDGVPEAISRG--LTTVSQPSLQKGRR
MUL_4146|M.ulcerans_Agy99 ALSAMDYLRGHGIYVPGQMTVTGFDGVPEAISRG--LTTVSQPSLQKGRR
MAV_0585|M.avium_104 ALSAMDYLRAHGVYVPGQLTVTGFDGVPDAISRG--LTTVSQSSLHKGRR
Mb3606c|M.bovis_AF2122/97 ALSAMDYLRAHGIYVPGQMTVTGFDGVPEALSRG--LTTVAQPSLHKGHR
Rv3575c|M.tuberculosis_H37Rv ALSAMDYLRAHGIYVPGQMTVTGFDGVPEALSRG--LTTVAQPSLHKGHR
Mflv_1457|M.gilvum_PYR-GCK ALSAMDHLRAKGIYVPGQMTVTGFDGVPEARQRG--LTTVAQPGVEKGRR
Mvan_2545|M.vanbaalenii_PYR-1 ALGALRTLRRSGISVPQQVSVIGVDDHPAAELSD--LTTVAQPVREQGRI
MSMEG_2636|M.smegmatis_MC2_155 AIGAIAAWRELGLSVPDDVGIAGFDDIPTLRDHTPALTTVVLPLQDIGVR
MAB_2023c|M.abscessus_ATCC_199 AARSLPVLQSLGHEVGSNFHLGTLVGDPATELTNPDIHAVDLRPREFGHQ
* :: : * * :. : . . * : :* . *
MMAR_5070|M.marinum_M AGELLQNPPRSG-----LPVVELLDTELIRGRTAGPPA-------
MUL_4146|M.ulcerans_Agy99 AGELLQNPPRSG-----LPVVELLDTELIRGRTAGPPA-------
MAV_0585|M.avium_104 AGELLLKPARSG-----LPVVELLDTELIRGRTAGPPA-------
Mb3606c|M.bovis_AF2122/97 AGELLLKPPRSG-----LPVIEVLDTELVRGRTAGPPA-------
Rv3575c|M.tuberculosis_H37Rv AGELLLKPPRSG-----LPVIEVLDTELVRGRTAGPPA-------
Mflv_1457|M.gilvum_PYR-GCK AGELLHNPPRSG-----LPVVEVLDTEVIRGRTSGPPA-------
Mvan_2545|M.vanbaalenii_PYR-1 AARVVLGGLGDGDASGPPASVTTLPTRLVVRGSTAPPR-------
MSMEG_2636|M.smegmatis_MC2_155 AVELALRTDADCD-----DLRERIPGRVVLRDSTRLP--------
MAB_2023c|M.abscessus_ATCC_199 AVTFLHDIIAGRAS---ANDRRTHQARLKIAGVSSPPAIRRAMSD
* . . .: : *