For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTTMRDETHAAGSQVRRAVLGDDHVDRAVAATTGFTADFQDLITRYAWGEIWTRPGLDRRSRSMITLTAM IARGHHDELAMHVRAARRNGLSVDEIKEVLLQSAIYCGVPEANTAFRIAGAVLAEEAQEPGDER
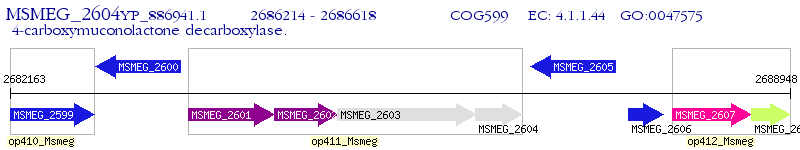
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2604 | pcaC | - | 100% (134) | 4-carboxymuconolactone decarboxylase |
| M. smegmatis MC2 155 | MSMEG_6150 | - | 2e-16 | 40.18% (112) | 4-carboxymuconolactone decarboxylase |
| M. smegmatis MC2 155 | MSMEG_5814 | pcaC | 1e-08 | 36.56% (93) | hypothetical protein MSMEG_5814 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0794 | - | 1e-08 | 36.49% (74) | 4-carboxymuconolactone decarboxylase |
| M. gilvum PYR-GCK | Mflv_0527 | - | 2e-50 | 73.81% (126) | 4-carboxymuconolactone decarboxylase |
| M. tuberculosis H37Rv | Rv0771 | - | 1e-08 | 36.49% (74) | 4-carboxymuconolactone decarboxylase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1221 | - | 4e-06 | 31.13% (106) | 4-carboxymuconolactone decarboxylase |
| M. marinum M | MMAR_4925 | - | 2e-09 | 42.25% (71) | 4-carboxymuconolactone decarboxylase |
| M. avium 104 | MAV_3552 | pcaC | 2e-11 | 32.00% (125) | hypothetical protein MAV_3552 |
| M. thermoresistible (build 8) | TH_0621 | - | 3e-18 | 49.41% (85) | 4-carboxymuconolactone decarboxylase |
| M. ulcerans Agy99 | MUL_0480 | - | 2e-09 | 42.25% (71) | 4-carboxymuconolactone decarboxylase |
| M. vanbaalenii PYR-1 | Mvan_0563 | - | 2e-50 | 73.02% (126) | 4-carboxymuconolactone decarboxylase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0794|M.bovis_AF2122/97 --------------------------------------------------
Rv0771|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4925|M.marinum_M --------------------------------------------------
MUL_0480|M.ulcerans_Agy99 --------------------------------------------------
MAB_1221|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0621|M.thermoresistible__bu --------------------------------------------------
Mflv_0527|M.gilvum_PYR-GCK MSNPPLTAIHLGGPDDGPLLLLGPSLGTTTATLWTGVAQRLVDHVRVVGW
Mvan_0563|M.vanbaalenii_PYR-1 MSNPPLTAIHLGGPDDGPLLLLGPSLGTTTATLWTGVAQRLVDHVRVVGW
MSMEG_2604|M.smegmatis_MC2_155 --------------------------------------------------
MAV_3552|M.avium_104 --------------------------------------------------
Mb0794|M.bovis_AF2122/97 --------------------------------------------------
Rv0771|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4925|M.marinum_M --------------------------------------------------
MUL_0480|M.ulcerans_Agy99 --------------------------------------------------
MAB_1221|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0621|M.thermoresistible__bu --------------------------------------------------
Mflv_0527|M.gilvum_PYR-GCK DLPGHGRGRRAHPFTIADLAAAVLVIADDLNVETFHYAGDSVGGCVGLQL
Mvan_0563|M.vanbaalenii_PYR-1 DLPGHGRGRRAHPFTIADLAAAVLVIADDLNVETFHYAGDSVGGCVGLQL
MSMEG_2604|M.smegmatis_MC2_155 --------------------------------------------------
MAV_3552|M.avium_104 --------------------------------------------------
Mb0794|M.bovis_AF2122/97 --------------------------------------------------
Rv0771|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4925|M.marinum_M --------------------------------------------------
MUL_0480|M.ulcerans_Agy99 --------------------------------------------------
MAB_1221|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0621|M.thermoresistible__bu --------------------------------------------------
Mflv_0527|M.gilvum_PYR-GCK LLDAPQRVSSATLLCTGAAIGTPDGWLARAATVRAGGVDTMLTGAAERWF
Mvan_0563|M.vanbaalenii_PYR-1 LLDAPQRVSSATLLCTGAAIGTPDGWLARAATVRAGGVDTMLAGAAERWF
MSMEG_2604|M.smegmatis_MC2_155 --------------------------------------------------
MAV_3552|M.avium_104 --------------------------------------------------
Mb0794|M.bovis_AF2122/97 --------------------------------------------------
Rv0771|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4925|M.marinum_M --------------------------------------------------
MUL_0480|M.ulcerans_Agy99 --------------------------------------------------
MAB_1221|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0621|M.thermoresistible__bu --------------------------------------------------
Mflv_0527|M.gilvum_PYR-GCK APGFVDREPGTASALLDALSHTDAESYAQVCEALAVFDVTDRLSEIVTPV
Mvan_0563|M.vanbaalenii_PYR-1 APGFVDREPGTASALLDALSHTDAESYAQVCEALAVFDVTNQLSEIVTPV
MSMEG_2604|M.smegmatis_MC2_155 --------------------------------------------------
MAV_3552|M.avium_104 --------------------------------------------------
Mb0794|M.bovis_AF2122/97 --------------------------------------------------
Rv0771|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4925|M.marinum_M --------------------------------------------------
MUL_0480|M.ulcerans_Agy99 --------------------------------------------------
MAB_1221|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0621|M.thermoresistible__bu --------------------------------------------------
Mflv_0527|M.gilvum_PYR-GCK LAVAGSADSPTPPESLRRIASDVKDGDVVVLEGVGHLAPAEAPERVAGLI
Mvan_0563|M.vanbaalenii_PYR-1 LAVAGSVDVPTPPESLRRIASDVKDGDLVVLEGVGHLAPAEAPQRVAGLI
MSMEG_2604|M.smegmatis_MC2_155 --------------------------------------------------
MAV_3552|M.avium_104 --------------------------------------------------
Mb0794|M.bovis_AF2122/97 ------------MMDELRRTGLDKMNEVYAWDMPD--------MPG-EFF
Rv0771|M.tuberculosis_H37Rv ------------MMDELRRTGLDKMNEVYAWDMPD--------MPG-EFF
MMAR_4925|M.marinum_M -------------MDELRRKGLDKMNEVYGWEMPN--------IEGDAYF
MUL_0480|M.ulcerans_Agy99 -------------MDELRRKGLDKMNEVYGWEMPN--------IEGDAYF
MAB_1221|M.abscessus_ATCC_1997 ---MVCHTRKGQVMDELRRQGLDKMKEVYGWDMPD--------IPG-DYF
TH_0621|M.thermoresistible__bu ----------------MREQGLQVFRELLPGVLPDGDVDFRDGSFGDELL
Mflv_0527|M.gilvum_PYR-GCK AEIVGVPQPPSKTLEDVHRAGMAVRREVLGHAHVDR-AVAGTTDLTADFQ
Mvan_0563|M.vanbaalenii_PYR-1 AEIVGVPQPPSKTLEDVHRAGMAVRREVLGEAHVDR-AVAGTTDLTADFQ
MSMEG_2604|M.smegmatis_MC2_155 ---------MTTMRDETHAAGSQVRRAVLGDDHVDR-AVAATTGFTADFQ
MAV_3552|M.avium_104 --------MSRSDSAARHQRGVRTFAEVMTFDPPDD---------ASPCA
: * : :
Mb0794|M.bovis_AF2122/97 ALTVDHLFGRIWTRPGLSMRDRRMAVIAVLTAQGQSDLLEVQVNAVLHND
Rv0771|M.tuberculosis_H37Rv ALTVDHLFGRIWTRPGLSMRDRRMAVIAVLTAQGQSDLLEVQVNAVLHND
MMAR_4925|M.marinum_M DLTVDHLFGTIWTRPGLSMRDRRLMTLTAVTAIGNRDLAEIQINAALLNG
MUL_0480|M.ulcerans_Agy99 DLTVDHLFGTIWTRPGLSMRDRRLMTLTAVTAIGNRDLAEIQINAAPLNG
MAB_1221|M.abscessus_ATCC_1997 KYTIDHLFGTIWTRPELSMRDRRLLLLGAVSALGLDEILEIQTSAALKNG
TH_0621|M.thermoresistible__bu EIGIDNVFGRLWTRDGLSRRDRSLVTLGILIALRATDELRFHVQIARNNG
Mflv_0527|M.gilvum_PYR-GCK HMITQYAWGSIWTRPGLDFRSRSMITLTALVARGHHEELAMHLRAARRNG
Mvan_0563|M.vanbaalenii_PYR-1 HMITQYAWGTIWTRPGLDLRSRSMITLTALVARGHHEELAMHLRAARRNG
MSMEG_2604|M.smegmatis_MC2_155 DLITRYAWGEIWTRPGLDRRSRSMITLTAMIARGHHDELAMHVRAARRNG
MAV_3552|M.avium_104 SGLIDFVFGEVWSRPGLSRRDRRFVTLACVAAADAQAPLEQHVYAALNSG
:* :*:* *. *.* : : : * : . ..
Mb0794|M.bovis_AF2122/97 ELTIDELRELAVFITHYVGFPLGSRLNSAIERVAAKRKQAAENGSLPDTK
Rv0771|M.tuberculosis_H37Rv ELTIDELRELAVFITHYVGFPLGSRLNSAIERVAAKRKQAAENGSLPDTK
MMAR_4925|M.marinum_M ELTEAELKEMAVFLTHYLGFPLGSGLNGAVDTVVARRKKAAAKGAGEDKK
MUL_0480|M.ulcerans_Agy99 ELTEAELKEMAVFLTHYLGFPLGSGLNGAVDTVVARRKKAAAKGAGEDKK
MAB_1221|M.abscessus_ATCC_1997 ELTEAELREVAIFITHYVGWPLGQRVNKHVEMAVAKQNEARR------TK
TH_0621|M.thermoresistible__bu -LTEDEIAEVIYHATGYAGFPNASTAMKVAKEVLAEEHSRAADADKQSRL
Mflv_0527|M.gilvum_PYR-GCK -LSNDEIKELLLQTAIYCGVPDANSAFRIAAEVLPEFDEHPGAPS-----
Mvan_0563|M.vanbaalenii_PYR-1 -LSNDEIKELLMQTAIYCGVPDANSAFRIAAEVLPEFDEHPGAPS-----
MSMEG_2604|M.smegmatis_MC2_155 -LSVDEIKEVLLQSAIYCGVPEANTAFRIAGAVLAEEAQEPGDER-----
MAV_3552|M.avium_104 DLSITEMREAVLHFAVYAGWPKASRFNIVVDQQWARIAAERGEPTPDPEP
*: *: * : * * * .. ..
Mb0794|M.bovis_AF2122/97 ANVAEVLAKESGKSS-----------------------------------
Rv0771|M.tuberculosis_H37Rv ANVAEVLAKESGKSS-----------------------------------
MMAR_4925|M.marinum_M GNVDDALKMHSGGKPD----------------------------------
MUL_0480|M.ulcerans_Agy99 GNVDDALKMHSGSKPD----------------------------------
MAB_1221|M.abscessus_ATCC_1997 ESSDD---------------------------------------------
TH_0621|M.thermoresistible__bu GESNP---------------------------------------------
Mflv_0527|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0563|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2604|M.smegmatis_MC2_155 --------------------------------------------------
MAV_3552|M.avium_104 LLPLVTAADPEQRLRGGERSFKDINCLPFAPPRDNPYSGAGILNFVFGEM
Mb0794|M.bovis_AF2122/97 --------------------------------------------------
Rv0771|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4925|M.marinum_M --------------------------------------------------
MUL_0480|M.ulcerans_Agy99 --------------------------------------------------
MAB_1221|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0621|M.thermoresistible__bu --------------------------------------------------
Mflv_0527|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0563|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2604|M.smegmatis_MC2_155 --------------------------------------------------
MAV_3552|M.avium_104 WLRPGLGMKERRLITVACVAFQDAEIPIMSHVYAALKSGDVSFREMDEVA
Mb0794|M.bovis_AF2122/97 --------------------------------------
Rv0771|M.tuberculosis_H37Rv --------------------------------------
MMAR_4925|M.marinum_M --------------------------------------
MUL_0480|M.ulcerans_Agy99 --------------------------------------
MAB_1221|M.abscessus_ATCC_1997 --------------------------------------
TH_0621|M.thermoresistible__bu --------------------------------------
Mflv_0527|M.gilvum_PYR-GCK --------------------------------------
Mvan_0563|M.vanbaalenii_PYR-1 --------------------------------------
MSMEG_2604|M.smegmatis_MC2_155 --------------------------------------
MAV_3552|M.avium_104 LQFAAYYGCAKADHLNQALAEQKQRVIAENHSDATPVG