For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAAVTRDADVQRVGFLGLGNMGRPMARRLVASGTPLTVYNRTAAKCAEFADLGCEVAESVARMGHCSVLL TMLGTDDDVAQVYLGDDGLVANARAGAVLVDCSTISPDMSARVRARCDRAGVHFLAAPVAGGPPVIESGG LAMAVSGDRQAFEHAADVLRVVAPNLIYVGPGDTSRLVKICHNLLVAATLEVLGELCVLAESHGASRQAL LSFLRSTAISSRFIEYKAPLLESLDFTPAFTSSLMQKDLELGLGLAGQADVTMPVTAQVHATYRAANESG LAELDAAAVYLYLAGSNPPERAGGGDDRSGRNGGPARPDGAPDPHR
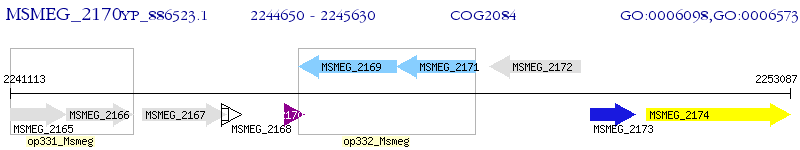
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2170 | - | - | 100% (326) | 3-hydroxyisobutyrate dehydrogenase family protein |
| M. smegmatis MC2 155 | MSMEG_5884 | - | 6e-35 | 34.56% (298) | 3-hydroxyisobutyrate dehydrogenase family protein |
| M. smegmatis MC2 155 | MSMEG_5477 | - | 1e-32 | 27.69% (307) | 2-hydroxy-3-oxopropionate reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0773c | mmsB | 1e-27 | 29.86% (288) | 3-hydroxyisobutyrate dehydrogenase MmsB |
| M. gilvum PYR-GCK | Mflv_2481 | - | 1e-30 | 32.00% (250) | 6-phosphogluconate dehydrogenase, NAD-binding |
| M. tuberculosis H37Rv | Rv0751c | mmsB | 1e-27 | 29.86% (288) | 3-hydroxyisobutyrate dehydrogenase MmsB |
| M. leprae Br4923 | MLBr_02065 | gnd | 2e-08 | 21.41% (327) | 6-phosphogluconate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1043c | - | 7e-28 | 29.75% (279) | putative oxidoreductase |
| M. marinum M | MMAR_1077 | mmsB | 8e-29 | 29.73% (296) | 3-hydroxyisobutyrate dehydrogenase, MmsB |
| M. avium 104 | MAV_4419 | mmsB | 9e-28 | 29.83% (295) | 3-hydroxyisobutyrate dehydrogenase |
| M. thermoresistible (build 8) | TH_2512 | - | 7e-32 | 33.33% (252) | PUTATIVE 6-phosphogluconate dehydrogenase, NAD-binding |
| M. ulcerans Agy99 | MUL_0835 | mmsB | 1e-28 | 29.69% (293) | 3-hydroxyisobutyrate dehydrogenase, MmsB |
| M. vanbaalenii PYR-1 | Mvan_5187 | - | 1e-32 | 31.56% (301) | 6-phosphogluconate dehydrogenase, NAD-binding |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2481|M.gilvum_PYR-GCK ----------MRVGFIGLGSQGGPMARRIAEGGFETTLWARRQASLEPYS
TH_2512|M.thermoresistible__bu ----------MRVGFIGLGSQGGPMARRIAENGYETTLWARRAATLEPFA
MAB_1043c|M.abscessus_ATCC_199 ---------MSTIAVIGLGAMGKAMADNLVQAGHQVSVWNRSPAPVAELA
Mvan_5187|M.vanbaalenii_PYR-1 ----------MKIGFIGLGNMGSAMAANLLSAGHEVLAYNRSRDRVVALA
Mb0773c|M.bovis_AF2122/97 ---------MTTIAFLGLGNMGAPMSANLVGAGHVVRGFDPAPTAASGAA
Rv0751c|M.tuberculosis_H37Rv ---------MTTIAFLGLGNMGAPMSANLVGAGHVVRGFDPAPTAASGAA
MMAR_1077|M.marinum_M ------MTKSCQIAFLGLGHMGAPMSANLIGAGHTVRGFDPVPAAASAAA
MUL_0835|M.ulcerans_Agy99 ------MTKSCQIAFLGLGHMGAPMSANLIGAGHTVRGFDPVPAAASATA
MAV_4419|M.avium_104 ----------MVVAFLGLGNMGAPMAANLVAAQHAVRGFDPVPAALSAAT
MSMEG_2170|M.smegmatis_MC2_155 MAAVTRDADVQRVGFLGLGNMGRPMARRLVASGTPLTVYNRTAAKCAEFA
MLBr_02065|M.leprae_Br4923 MSAPESKTAIAQIGVTGLAVMGSNIARNFARHGYTVALHNRSIAKTDTLL
:.. **. * :: .:
Mflv_2481|M.gilvum_PYR-GCK DTP--------AKTASTPAELGAASDLVCLCVVGDDDVREVLYGEQGVLA
TH_2512|M.thermoresistible__bu DTP--------AKTAGSPAELGAASDLVCLCVVGDDDVREVLYGDTGVLA
MAB_1043c|M.abscessus_ATCC_199 AVG--------ARPLDSVAQA-FDSGVVFSVLADDHAVTETLL--TPEVL
Mvan_5187|M.vanbaalenii_PYR-1 ERG--------ATPVPTVAAA-CEGDVVFTMLADDTAVESVTFGDTGIVA
Mb0773c|M.bovis_AF2122/97 AHG--------VAVFRSAPEA-VAEADVVITMLPTGEVVRRCY--TDVLA
Rv0751c|M.tuberculosis_H37Rv AHG--------VAVFRSAPEA-VAEADVVITMLPTGEVVRRCY--TDVLA
MMAR_1077|M.marinum_M AKG--------ATMFDSAAEA-VAGADVVITMLPNGAVVKRCY--AEVLP
MUL_0835|M.ulcerans_Agy99 AKG--------ATMFDSAAEA-VARADVVITMLPNGAVVKRCY--AEVLP
MAV_4419|M.avium_104 ASG--------VTGFDTATDA-VTGADVVITMLPNGEVVKRCY--AEILP
MSMEG_2170|M.smegmatis_MC2_155 DLG--------CEVAESVARM-GHCSVLLTMLGTDDDVAQVYLGDDGLVA
MLBr_02065|M.leprae_Br4923 KEHGSEGNFVRTETIPEFLAALQTPRRVLIMVKAGDATDAVIN---ELAD
: : .
Mflv_2481|M.gilvum_PYR-GCK GLAEGGIIAIHSTVHPQTCQEIAEKGAAQGVSVIDAPVSGGGPAVEQGTL
TH_2512|M.thermoresistible__bu GLAPGGVVAIHSTVHPDTCREISEKAAAQQVSVIDAPVSGGGPAVEEGKL
MAB_1043c|M.abscessus_ATCC_199 AAGHGALHVNLATVSARLADRATALHDEHGVGYVAAPVFGRVPVAQAGQL
Mvan_5187|M.vanbaalenii_PYR-1 ALAPGAIHVSASTISVALSEKLTAAHAEAGQMFVSAPVFGRPEAAAAAKL
Mb0773c|M.bovis_AF2122/97 AARPATLFIDSSTISVTDAREVHALAESHGMLQLDAPVSGGVKGAAAATL
Rv0751c|M.tuberculosis_H37Rv AARPATLFIDSSTISVTDAREVHALAESHGMLQLDAPVSGGVKGAAAATL
MMAR_1077|M.marinum_M AARSGALFIDSSTISVNDAREVHALAESHGMAQLDAPVSGGVKGAVAGTL
MUL_0835|M.ulcerans_Agy99 AARSGALFIDSSTISVNDAREVHALAESHGMAQLDAPVSGGVKGAVAGTL
MAV_4419|M.avium_104 AARPGALFIDSSTISVADAREVHAQAQSHGVAQLDAPVSGGVKGAVAGTL
MSMEG_2170|M.smegmatis_MC2_155 NARAGAVLVDCSTISPDMSARVRARCDRAGVHFLAAPVAGGPPVIESGGL
MLBr_02065|M.leprae_Br4923 VMEPSDIIIDGGNSLFTDTIRREKAMRERGLHFVGAGISGGEEGALNG-P
. : .. . : * : * .
Mflv_2481|M.gilvum_PYR-GCK LVMVGGDEEVAERCRPVFATYADPIVHLG------PLGSGQNTKILNNLL
TH_2512|M.thermoresistible__bu LVMVGGDEEVVERCRPVFASYADPIVHLG------PLGSGQVTKILNNLL
MAB_1043c|M.abscessus_ATCC_199 QVLAAGAAAQIDKAQSFFDVIGARTWRLGD-----APRQANVVKIIGNFL
Mvan_5187|M.vanbaalenii_PYR-1 FVVAAGPHDAVTSLQPAFEAIGQRTYVMSE-----EPKTANLVKLSGNFL
Mb0773c|M.bovis_AF2122/97 AFMVGGDESTLRRARPVLEPMAGKIIHCG------AAGAGQAAKVCNNMV
Rv0751c|M.tuberculosis_H37Rv AFMVGGDESTLRRARPVLEPMAGKIIHCG------AAGAGQAAKVCNNMV
MMAR_1077|M.marinum_M AFMVGGDQSALAQARPVLEPMAGKIIHCG------AAGAGQAAKVCNNMV
MUL_0835|M.ulcerans_Agy99 AFMVGGDQSALAQARPVLEPMAGKIIHCG------AAGAGQAAKVCNNMV
MAV_4419|M.avium_104 AFMVGGEPEALERARPVLEPMAGKIIHCG------AAGAGQAAKVCNNMV
MSMEG_2170|M.smegmatis_MC2_155 AMAVSGDRQAFEHAADVLRVVAPNLIYVG------PGDTSRLVKICHNLL
MLBr_02065|M.leprae_Br4923 SIMPGGPAESYTSLGPLLEEISAHVDGVSCCTHIGPGGSGHFVKMVHNGI
. .* : . . .. .*: * :
Mflv_2481|M.gilvum_PYR-GCK FSANLGSAVSTLELGES-LGIPRAKLVEVLSKG-----------------
TH_2512|M.thermoresistible__bu FTANLGSALSTLELGES-LGIPRVKLAEVLNGG-----------------
MAB_1043c|M.abscessus_ATCC_199 IASAIQSLSEAVSMAER-SGVDSALLVELLTST-----------------
Mvan_5187|M.vanbaalenii_PYR-1 IASVIESLGEAMALVSK-AGVDRQTYLELLTST-----------------
Mb0773c|M.bovis_AF2122/97 LAVQQIAIAEAFVLAEK-LGLSAQSLFDVITGA-----------------
Rv0751c|M.tuberculosis_H37Rv LAVQQIAIAEAFVLAEK-LGLSAQSLFDVITGA-----------------
MMAR_1077|M.marinum_M LAVQQIAIGEAFVLAEN-LGLDAQSLFDVITGA-----------------
MUL_0835|M.ulcerans_Agy99 LAVQQIAIGEAFVLAEN-LGLDAQSLFDVITGA-----------------
MAV_4419|M.avium_104 LAVQQIAIGEAFILAEK-LGLSAQSLFDVITGA-----------------
MSMEG_2170|M.smegmatis_MC2_155 VAATLEVLGELCVLAES-HGASRQALLSFLRST-----------------
MLBr_02065|M.leprae_Br4923 EYSDMQLIGEAYQLLRDGLGMSAPQIADVFTEWNRGDLNSYLVEITAEVL
. : * ..:
Mflv_2481|M.gilvum_PYR-GCK ---------------------SATSKAVSSIS-MFGGTLDGLAP------
TH_2512|M.thermoresistible__bu ---------------------SATSKALGSIS-VFGGTVEGLAP------
MAB_1043c|M.abscessus_ATCC_199 ---------------------LFQGPAYSSYGKLIATSAYEPAG------
Mvan_5187|M.vanbaalenii_PYR-1 ---------------------LFAAPAYQTYGGLLARKQFEPAG------
Mb0773c|M.bovis_AF2122/97 ---------------------TGNCWAVHTNCPVPGPVPTSPANNDFKPG
Rv0751c|M.tuberculosis_H37Rv ---------------------TGNCWAVHTNCPVPGPVPTSPANNDFKPG
MMAR_1077|M.marinum_M ---------------------TGNCWAVHTNCPVPGPVPTSPANNDFKPG
MUL_0835|M.ulcerans_Agy99 ---------------------TGNCWAVHTNCPVPGPVPTSPANNDFKPG
MAV_4419|M.avium_104 ---------------------TGNCWAVHTNCPVPGPVPTSPANNDFKPG
MSMEG_2170|M.smegmatis_MC2_155 ---------------------AISSRFIEYKAPLLESLDFTPAF------
MLBr_02065|M.leprae_Br4923 RQTDIKTGRPLVDVILDKAEQKGTGRWTVQSALDLGVPITGIAEAVFARA
*
Mflv_2481|M.gilvum_PYR-GCK IAGGLLQKDVRHAASLAAA--AKAPEGAVFTAADAALEAMDHRR------
TH_2512|M.thermoresistible__bu IAGALLQKDVRHAANIAAS--ASVPEGTVFTVADTALTAMGHARR-----
MAB_1043c|M.abscessus_ATCC_199 FTTTLGRKDVGLALEVAADTGLRLPFGEILRTVFDEALAGGHANLDWSSI
Mvan_5187|M.vanbaalenii_PYR-1 FAAHLGLKDVRLVLAAGEQLEVPLPVASLIRDRFLTLLAGGGRDLDWSAL
Mb0773c|M.bovis_AF2122/97 FSTALMNKDLGLAMDAVAATGATAPLGSHAADIYAKFAAD-HADLDFSAV
Rv0751c|M.tuberculosis_H37Rv FSTALMNKDLGLAMDAVAATGATAPLGSHAADIYAKFAAD-HADLDFSAV
MMAR_1077|M.marinum_M FATALMNKDLGLAMDAVTSTGSAAPLGSHAAEIYAKFAAD-HADQDFSAV
MUL_0835|M.ulcerans_Agy99 FATALMNKDLGLAMDAVTSTASAAPLGSHAAEIYAKFATD-HADQDFSAV
MAV_4419|M.avium_104 FAAALMNKDLGLAMDAVTSTGAAAPLGRHAAEIYAKFAAS-HGDKDFSAV
MSMEG_2170|M.smegmatis_MC2_155 -TSSLMQKDLELGLGLAGQADVTMPVTAQVHATYRAANESGLAELDAAAV
MLBr_02065|M.leprae_Br4923 LSGSVLQRKAAIGLASGKLGNKPTDRETFIEDVRQALYASKIVAYAQGFN
: : :.
Mflv_2481|M.gilvum_PYR-GCK --------------------------------------------------
TH_2512|M.thermoresistible__bu --------------------------------------------------
MAB_1043c|M.abscessus_ATCC_199 ADLQRARDAE----------------------------------------
Mvan_5187|M.vanbaalenii_PYR-1 GALADWEAGGPRPGSG----------------------------------
Mb0773c|M.bovis_AF2122/97 IHTLRARADA----------------------------------------
Rv0751c|M.tuberculosis_H37Rv IHTLRARADA----------------------------------------
MMAR_1077|M.marinum_M IEMLRGS-------------------------------------------
MUL_0835|M.ulcerans_Agy99 IEMLRGS-------------------------------------------
MAV_4419|M.avium_104 IEMLRGAC------------------------------------------
MSMEG_2170|M.smegmatis_MC2_155 YLYLAGSNPPERAGGGDDRSGRNGGPARPDGAPDPHR-------------
MLBr_02065|M.leprae_Br4923 HIQTGSTEFGWNITPGDLATIWRGGCIIRAKFLNRIKEAFDAAPDLASLI
Mflv_2481|M.gilvum_PYR-GCK --------------------------------------------------
TH_2512|M.thermoresistible__bu --------------------------------------------------
MAB_1043c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_5187|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb0773c|M.bovis_AF2122/97 --------------------------------------------------
Rv0751c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1077|M.marinum_M --------------------------------------------------
MUL_0835|M.ulcerans_Agy99 --------------------------------------------------
MAV_4419|M.avium_104 --------------------------------------------------
MSMEG_2170|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_02065|M.leprae_Br4923 VAPYFRSAVESAIDSWRRVVSTATQLGIPNPGFSSALSYYDALRTERLPA
Mflv_2481|M.gilvum_PYR-GCK -------------------------------------
TH_2512|M.thermoresistible__bu -------------------------------------
MAB_1043c|M.abscessus_ATCC_199 -------------------------------------
Mvan_5187|M.vanbaalenii_PYR-1 -------------------------------------
Mb0773c|M.bovis_AF2122/97 -------------------------------------
Rv0751c|M.tuberculosis_H37Rv -------------------------------------
MMAR_1077|M.marinum_M -------------------------------------
MUL_0835|M.ulcerans_Agy99 -------------------------------------
MAV_4419|M.avium_104 -------------------------------------
MSMEG_2170|M.smegmatis_MC2_155 -------------------------------------
MLBr_02065|M.leprae_Br4923 ALTQAQRDFFGAHSYGRVDARGKFHTLWSEDRSEVTV