For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
STIAFIGLGIMGSPMAVNLVKAGHTVKGYNRSPHRTADLVAAGGTAADSIADAVDGAEIVAVMVPDSPDV TEVLTADGGVFASAAPGTLIIDFSSIRPDVSMDLAKQAADHDLRLIDAPVSGGEIGAKNAALSIMIGGAD EDVSRAMPVLEAVGKTIVHVGPNGAGQTVKAANQLIVAGNIALLAEAIIFLEAYGVDTTAAVKVLGGGLA GSAVLDQKAQKMLDRSFDPGFRIELHHKDLGILTNAAREAGVVTPIGAAVAQLMASARINGDGALDHSGL FRTIAKLSGASTEPGSSNTKERP*
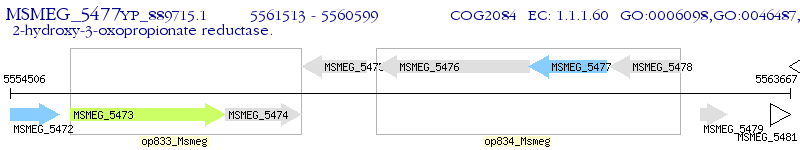
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5477 | - | - | 100% (304) | 2-hydroxy-3-oxopropionate reductase |
| M. smegmatis MC2 155 | MSMEG_1496 | mmsB | 5e-38 | 34.43% (273) | 3-hydroxyisobutyrate dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2170 | - | 1e-32 | 27.69% (307) | 3-hydroxyisobutyrate dehydrogenase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0773c | mmsB | 3e-41 | 34.81% (293) | 3-hydroxyisobutyrate dehydrogenase MmsB |
| M. gilvum PYR-GCK | Mflv_5012 | - | 6e-40 | 36.24% (287) | 3-hydroxyisobutyrate dehydrogenase |
| M. tuberculosis H37Rv | Rv0751c | mmsB | 3e-41 | 34.81% (293) | 3-hydroxyisobutyrate dehydrogenase MmsB |
| M. leprae Br4923 | MLBr_02065 | gnd | 4e-13 | 27.36% (201) | 6-phosphogluconate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1186c | - | 2e-40 | 35.74% (305) | 3-hydroxyisobutyrate dehydrogenase |
| M. marinum M | MMAR_1077 | mmsB | 2e-40 | 34.44% (270) | 3-hydroxyisobutyrate dehydrogenase, MmsB |
| M. avium 104 | MAV_4419 | mmsB | 5e-41 | 34.89% (278) | 3-hydroxyisobutyrate dehydrogenase |
| M. thermoresistible (build 8) | TH_0328 | mmsB | 9e-37 | 34.07% (273) | PROBABLE 3-HYDROXYISOBUTYRATE DEHYDROGENASE MMSB (HIBADH) |
| M. ulcerans Agy99 | MUL_0835 | mmsB | 5e-39 | 33.70% (270) | 3-hydroxyisobutyrate dehydrogenase, MmsB |
| M. vanbaalenii PYR-1 | Mvan_1360 | - | 2e-37 | 35.53% (273) | 3-hydroxyisobutyrate dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0773c|M.bovis_AF2122/97 ------MTT---IAFLGLGNMGAPMSANLVGAGHVVRGFDPAPTAASGAA
Rv0751c|M.tuberculosis_H37Rv ------MTT---IAFLGLGNMGAPMSANLVGAGHVVRGFDPAPTAASGAA
MMAR_1077|M.marinum_M ------MTKSCQIAFLGLGHMGAPMSANLIGAGHTVRGFDPVPAAASAAA
MUL_0835|M.ulcerans_Agy99 ------MTKSCQIAFLGLGHMGAPMSANLIGAGHTVRGFDPVPAAASATA
MAV_4419|M.avium_104 ----------MVVAFLGLGNMGAPMAANLVAAQHAVRGFDPVPAALSAAT
TH_0328|M.thermoresistible__bu --------------------MGGPMAANLVAAGHTVRGFDPVAESRAAAA
Mflv_5012|M.gilvum_PYR-GCK ------MTT---IAFLGLGNMGGPMAANLVAAGHTVHAFDVVPELRDAAE
Mvan_1360|M.vanbaalenii_PYR-1 ------MTT---IAFLGLGNMGGPMAANLVAAGQTVHAFDLVTELKEAAK
MAB_1186c|M.abscessus_ATCC_199 ------MTN---IAFIGLGHMGLPMAVNLTKAGHTVAAFDLSEQARDAAT
MSMEG_5477|M.smegmatis_MC2_155 ------MST---IAFIGLGIMGSPMAVNLVKAGHTVKGYNRSPHRTADLV
MLBr_02065|M.leprae_Br4923 MSAPESKTAIAQIGVTGLAVMGSNIARNFARHGYTVALHNRSIAKTDTLL
** :: *: .* .:
Mb0773c|M.bovis_AF2122/97 AHG--------VAVFRSAPEAVAEADVVITMLPTG---EVVRRCYTDVLA
Rv0751c|M.tuberculosis_H37Rv AHG--------VAVFRSAPEAVAEADVVITMLPTG---EVVRRCYTDVLA
MMAR_1077|M.marinum_M AKG--------ATMFDSAAEAVAGADVVITMLPNG---AVVKRCYAEVLP
MUL_0835|M.ulcerans_Agy99 AKG--------ATMFDSAAEAVARADVVITMLPNG---AVVKRCYAEVLP
MAV_4419|M.avium_104 ASG--------VTGFDTATDAVTGADVVITMLPNG---EVVKRCYAEILP
TH_0328|M.thermoresistible__bu DKG--------VQTFDSGAEAVAGAEVVITSLPNG---DIVKQCYAEVLP
Mflv_5012|M.gilvum_PYR-GCK AKG--------ANVFDSAAAAVADADVVVTSLPNG---AVVKAVYDEVVP
Mvan_1360|M.vanbaalenii_PYR-1 AKG--------AKVFDSGAAAVADADVVITSLPNG---NIVKAVYDEVVP
MAB_1186c|M.abscessus_ATCC_199 QAG--------VPLAESGAAAASTADVVITMLPKG---AHVLAAYEDLLP
MSMEG_5477|M.smegmatis_MC2_155 AAG--------GTAADSIADAVDGAEIVAVMVPDSPDVTEVLTADGGVFA
MLBr_02065|M.leprae_Br4923 KEHGSEGNFVRTETIPEFLAALQTPRRVLIMVKAG---DATDAVINELAD
* . * : . . :
Mb0773c|M.bovis_AF2122/97 AARPATLFIDSSTISVTDAREVHALAESHGMLQLDAPVSGGVKGAAAATL
Rv0751c|M.tuberculosis_H37Rv AARPATLFIDSSTISVTDAREVHALAESHGMLQLDAPVSGGVKGAAAATL
MMAR_1077|M.marinum_M AARSGALFIDSSTISVNDAREVHALAESHGMAQLDAPVSGGVKGAVAGTL
MUL_0835|M.ulcerans_Agy99 AARSGALFIDSSTISVNDAREVHALAESHGMAQLDAPVSGGVKGAVAGTL
MAV_4419|M.avium_104 AARPGALFIDSSTISVADAREVHAQAQSHGVAQLDAPVSGGVKGAVAGTL
TH_0328|M.thermoresistible__bu AATSGALFIDTSTISVDDARTVHAQATGRGFAQVDAPVSGGVKGAVAGTL
Mflv_5012|M.gilvum_PYR-GCK AAKAGALLIDTSTISVDDARAIHEQATAGGLAQIDAPVSGGIKGATAGTL
Mvan_1360|M.vanbaalenii_PYR-1 AAKAGALLIDTSTISVDDARAIHDQATEKGLAQIDAPVSGGIKGATAGTL
MAB_1186c|M.abscessus_ATCC_199 AARPGTLFIDSSTVDVADARAAHEAATAAGHRFADAPVSGGVPGATAATL
MSMEG_5477|M.smegmatis_MC2_155 SAAPGTLIIDFSSIRPDVSMDLAKQAADHDLRLIDAPVSGGEIGAKNAAL
MLBr_02065|M.leprae_Br4923 VMEPSDIIIDGGNSLFTDTIRREKAMRERGLHFVGAGISGGEEGALNG-P
.. ::** .. : . .* :*** ** .
Mb0773c|M.bovis_AF2122/97 AFMVGGDESTLRRARPVLEPMAGKII------HCGAAGAGQAAKVCNNMV
Rv0751c|M.tuberculosis_H37Rv AFMVGGDESTLRRARPVLEPMAGKII------HCGAAGAGQAAKVCNNMV
MMAR_1077|M.marinum_M AFMVGGDQSALAQARPVLEPMAGKII------HCGAAGAGQAAKVCNNMV
MUL_0835|M.ulcerans_Agy99 AFMVGGDQSALAQARPVLEPMAGKII------HCGAAGAGQAAKVCNNMV
MAV_4419|M.avium_104 AFMVGGEPEALERARPVLEPMAGKII------HCGAAGAGQAAKVCNNMV
TH_0328|M.thermoresistible__bu AFMVGGDAEAVERARPVLEPMAGKII------HCGASGTGQAAKLCNNMV
Mflv_5012|M.gilvum_PYR-GCK AFMVGGDDEAFERAKPVLEPMAGKII------HCGASGAGQAAKLCNNMV
Mvan_1360|M.vanbaalenii_PYR-1 AFMVGGEDEAFERAKPVLEPMAGKII------HCGASGAGQAAKLCNNMV
MAB_1186c|M.abscessus_ATCC_199 TFMVGADDAVFVAAEPILDAMGKRVV------HCGGPGVGQAAKICNNMI
MSMEG_5477|M.smegmatis_MC2_155 SIMIGGADEDVSRAMPVLEAVGKTIV------HVGPNGAGQTVKAANQLI
MLBr_02065|M.leprae_Br4923 SIMPGGPAESYTSLGPLLEEISAHVDGVSCCTHIGPGGSGHFVKMVHNGI
::* *. *:*: :. : * * * *: .* :: :
Mb0773c|M.bovis_AF2122/97 LAVQQIAIAEAFVLAEK-LGLSAQSLFDVITGATG---NCWAVHTNCPVP
Rv0751c|M.tuberculosis_H37Rv LAVQQIAIAEAFVLAEK-LGLSAQSLFDVITGATG---NCWAVHTNCPVP
MMAR_1077|M.marinum_M LAVQQIAIGEAFVLAEN-LGLDAQSLFDVITGATG---NCWAVHTNCPVP
MUL_0835|M.ulcerans_Agy99 LAVQQIAIGEAFVLAEN-LGLDAQSLFDVITGATG---NCWAVHTNCPVP
MAV_4419|M.avium_104 LAVQQIAIGEAFILAEK-LGLSAQSLFDVITGATG---NCWAVHTNCPVP
TH_0328|M.thermoresistible__bu LAVQQIAIGEAFVLAEK-LGLSAQSLFDVITGATG---NCWAVHTNCPVP
Mflv_5012|M.gilvum_PYR-GCK LAVQQIAIGEAFVLAEK-LGLPAQSLFDVITGATG---NCWSVHTNCPVP
Mvan_1360|M.vanbaalenii_PYR-1 LAVQQIAIGEAFVLAEK-LGLPAQSLFDVITGATG---NCWAVHTNCPVP
MAB_1186c|M.abscessus_ATCC_199 LGVSMIAISEAFVLGEK-LGLSHQALFDVAANASG---QCWALTSNCPVP
MSMEG_5477|M.smegmatis_MC2_155 VAGNIALLAEAIIFLEA-YGVDTTAAVKVLGGG----------LAGSAVL
MLBr_02065|M.leprae_Br4923 EYSDMQLIGEAYQLLRDGLGMSAPQIADVFTEWNRGDLNSYLVEITAEVL
. :.** : . *: .* . *
Mb0773c|M.bovis_AF2122/97 GPVPTSPAN----------NDFKPGFSTALMNKDLGLAMDAVAATGATAP
Rv0751c|M.tuberculosis_H37Rv GPVPTSPAN----------NDFKPGFSTALMNKDLGLAMDAVAATGATAP
MMAR_1077|M.marinum_M GPVPTSPAN----------NDFKPGFATALMNKDLGLAMDAVTSTGSAAP
MUL_0835|M.ulcerans_Agy99 GPVPTSPAN----------NDFKPGFATALMNKDLGLAMDAVTSTASAAP
MAV_4419|M.avium_104 GPVPTSPAN----------NDFKPGFAAALMNKDLGLAMDAVTSTGAAAP
TH_0328|M.thermoresistible__bu GPVPTSPAN----------NDFKPGFATALMNKDLGLAMDAVKSSGASAP
Mflv_5012|M.gilvum_PYR-GCK GPVPTSPAN----------NDFKPGFATALMNKDLGLAMAAVESSGAKAP
Mvan_1360|M.vanbaalenii_PYR-1 GPVPTSPAN----------NDFKPGFATALMNKDLGLAMAAVSSTGSSAP
MAB_1186c|M.abscessus_ATCC_199 GPVPTSPAN----------RDYAPGFAVALMAKDLGLAANAVRANGVDAQ
MSMEG_5477|M.smegmatis_MC2_155 DQKAQKMLD----------RSFDPGFRIELHHKDLGILTNAAREAGVVTP
MLBr_02065|M.leprae_Br4923 RQTDIKTGRPLVDVILDKAEQKGTGRWTVQSALDLGVPITGIAEAVFARA
. .. .* ***: .
Mb0773c|M.bovis_AF2122/97 LGSHAADIYAKF----AAD-HADLDFSAVIHTLRARADA-----------
Rv0751c|M.tuberculosis_H37Rv LGSHAADIYAKF----AAD-HADLDFSAVIHTLRARADA-----------
MMAR_1077|M.marinum_M LGSHAAEIYAKF----AAD-HADQDFSAVIEMLRGS--------------
MUL_0835|M.ulcerans_Agy99 LGSHAAEIYAKF----ATD-HADQDFSAVIEMLRGS--------------
MAV_4419|M.avium_104 LGRHAAEIYAKF----AAS-HGDKDFSAVIEMLRGAC-------------
TH_0328|M.thermoresistible__bu LGSHAAEIYAEFIR--AADGNADKDFSAVIELLRNS--------------
Mflv_5012|M.gilvum_PYR-GCK LGTHAAEIYSDF-----AKEHAGKDFSAIIEALRD---------------
Mvan_1360|M.vanbaalenii_PYR-1 LGTHAAEIYAEF-----NKEHADKDFSAIIEALRG---------------
MAB_1186c|M.abscessus_ATCC_199 LGLAAAHIYDDFN----STGGSGLDFSAIFSRIQGAS-------------
MSMEG_5477|M.smegmatis_MC2_155 IGAAVAQLMASAR----INGDGALDHSGLFRTIAKLSGASTEPGSSNTKE
MLBr_02065|M.leprae_Br4923 LSGSVLQRKAAIGLASGKLGNKPTDRETFIEDVRQALYASKIVAYAQGFN
:. . . * . .: :
Mb0773c|M.bovis_AF2122/97 --------------------------------------------------
Rv0751c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1077|M.marinum_M --------------------------------------------------
MUL_0835|M.ulcerans_Agy99 --------------------------------------------------
MAV_4419|M.avium_104 --------------------------------------------------
TH_0328|M.thermoresistible__bu --------------------------------------------------
Mflv_5012|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1360|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_1186c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5477|M.smegmatis_MC2_155 RP------------------------------------------------
MLBr_02065|M.leprae_Br4923 HIQTGSTEFGWNITPGDLATIWRGGCIIRAKFLNRIKEAFDAAPDLASLI
Mb0773c|M.bovis_AF2122/97 --------------------------------------------------
Rv0751c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1077|M.marinum_M --------------------------------------------------
MUL_0835|M.ulcerans_Agy99 --------------------------------------------------
MAV_4419|M.avium_104 --------------------------------------------------
TH_0328|M.thermoresistible__bu --------------------------------------------------
Mflv_5012|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1360|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_1186c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5477|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_02065|M.leprae_Br4923 VAPYFRSAVESAIDSWRRVVSTATQLGIPNPGFSSALSYYDALRTERLPA
Mb0773c|M.bovis_AF2122/97 -------------------------------------
Rv0751c|M.tuberculosis_H37Rv -------------------------------------
MMAR_1077|M.marinum_M -------------------------------------
MUL_0835|M.ulcerans_Agy99 -------------------------------------
MAV_4419|M.avium_104 -------------------------------------
TH_0328|M.thermoresistible__bu -------------------------------------
Mflv_5012|M.gilvum_PYR-GCK -------------------------------------
Mvan_1360|M.vanbaalenii_PYR-1 -------------------------------------
MAB_1186c|M.abscessus_ATCC_199 -------------------------------------
MSMEG_5477|M.smegmatis_MC2_155 -------------------------------------
MLBr_02065|M.leprae_Br4923 ALTQAQRDFFGAHSYGRVDARGKFHTLWSEDRSEVTV