For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
EIDFTRLRYFVAVAEELHFKRAADRLMITPPPLSKQIKLLEKELGGQLFERNYHEVRLTPLGEKLLGPAR DILARVEDLKATASRITQGVAPVRVGATAYAPSDFLARLEKAVAGLAAPTQFSVPGSAAEVTAKLVSGHL DIGLIHLPSADKRLRHRVVARYQGAIAVRFDDPLAAKDVVAIEELRDREVVIDFARPNPVVLAGLTRGLN AKGVTKIVRAINQLGGELEMATQVFNRHLVAMVTYAPESMIGRIFSPPEFKLIPIDEATWAPAQIALAWA PDRLSDRRSERVLRPAEIEALVDELERAVARPAGGA*
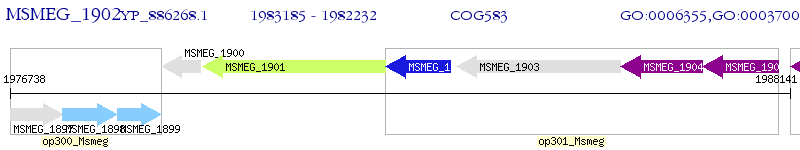
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1902 | - | - | 100% (317) | transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_1895 | - | 1e-24 | 35.90% (195) | HTH-type transcriptional regulator AlsR |
| M. smegmatis MC2 155 | MSMEG_1909 | - | 8e-21 | 29.39% (330) | putative HTH-type transcriptional regulator YnfL |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0121 | oxyS | 6e-12 | 33.33% (150) | oxidative stress response regulatory protein OXYS |
| M. gilvum PYR-GCK | Mflv_2332 | - | 1e-129 | 74.18% (306) | LysR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0117 | oxyS | 6e-12 | 33.33% (150) | oxidative stress response regulatory protein OXYS |
| M. leprae Br4923 | MLBr_02663 | oxyS | 2e-13 | 41.41% (99) | putative LysR family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_3208c | - | 4e-20 | 32.29% (192) | LysR family transcriptional regulator |
| M. marinum M | MMAR_0317 | oxyS | 2e-12 | 32.00% (150) | oxidative stress response regulatory protein OxyS |
| M. avium 104 | MAV_3102 | - | 4e-14 | 32.92% (161) | LysR substrate binding domain-containing protein |
| M. thermoresistible (build 8) | TH_3504 | - | 1e-13 | 26.86% (283) | PUTATIVE putative transcriptional regulator |
| M. ulcerans Agy99 | MUL_4805 | oxyS | 3e-12 | 39.39% (99) | oxidative stress response regulatory protein OxyS |
| M. vanbaalenii PYR-1 | Mvan_4315 | - | 1e-129 | 73.86% (306) | LysR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2332|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4315|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1902|M.smegmatis_MC2_155 --------------------------------------------------
Mb0121|M.bovis_AF2122/97 --------------------------------------------------
Rv0117|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_02663|M.leprae_Br4923 --------------------------------------------------
MMAR_0317|M.marinum_M MKLLGAPKINPELTLAQIPGSDSLVVIPIPRTSGLARRAAPAVQPRCGVT
MUL_4805|M.ulcerans_Agy99 --------------------------------------------------
MAB_3208c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3102|M.avium_104 --------------------------------------------------
TH_3504|M.thermoresistible__bu --------------------------------------------------
Mflv_2332|M.gilvum_PYR-GCK -------------------------------------------MDIDFTR
Mvan_4315|M.vanbaalenii_PYR-1 -------------------------------------------MDIDFTR
MSMEG_1902|M.smegmatis_MC2_155 -------------------------------------------MEIDFTR
Mb0121|M.bovis_AF2122/97 ---------------------------------------------MLFRQ
Rv0117|M.tuberculosis_H37Rv ---------------------------------------------MLFRQ
MLBr_02663|M.leprae_Br4923 ---------------------------------------------MLFRQ
MMAR_0317|M.marinum_M RTAQWRADLVATVQDLAAQSGTGGRQRSGGGGSLSESVPMTEEVSVLFRQ
MUL_4805|M.ulcerans_Agy99 ---------------------------------------------MLFRQ
MAB_3208c|M.abscessus_ATCC_199 ---------------------------------------------MELRQ
MAV_3102|M.avium_104 ---------------------------------------------MDTHR
TH_3504|M.thermoresistible__bu ----------------------------------------VPSPKIDLRR
: :
Mflv_2332|M.gilvum_PYR-GCK LRYFVAVADELHFKRAADKLRITPPPLSKQIKLLEKELGGPLFERGYHEV
Mvan_4315|M.vanbaalenii_PYR-1 LRYFVAVADELHFKRAADKLMITPPPLSKQIKLLEKELGGPLFERGYHDV
MSMEG_1902|M.smegmatis_MC2_155 LRYFVAVAEELHFKRAADRLMITPPPLSKQIKLLEKELGGQLFERNYHEV
Mb0121|M.bovis_AF2122/97 LEYFVAVAQERHFARAAEKCYVSQPALSSAIAKLERELNVTLINRGHSFE
Rv0117|M.tuberculosis_H37Rv LEYFVAVAQERHFARAAEKCYVSQPALSSAIAKLERELNVTLINRGHSFE
MLBr_02663|M.leprae_Br4923 LEYFVAVAQERHFARAAERCYVSQPALSSAIAKLERELNVTLINRGHSFE
MMAR_0317|M.marinum_M LEYFVAVAQERHFARAAEKCYVSQPALSSAIAKLERELNVTLINRGHSFE
MUL_4805|M.ulcerans_Agy99 LEYFVAVAQERHFARAAEKCYVSQPALSSAIAKLERELHVTLINRGHSFE
MAB_3208c|M.abscessus_ATCC_199 LEYFLAVAEHANFTRAAEALHVAQPWVSAQVRRLERELGNELFDRSSRVL
MAV_3102|M.avium_104 LKYFLRIAEQGSITGAAASLGIAQPALSRQIRLLEEDLGVALFRRTRRGV
TH_3504|M.thermoresistible__bu LAQFVAVAELGSFTRAAATLHLSQQALSSAVRALENDVGADLFTRDGRRI
* *: :*: : ** :: :* : **.:: *: *
Mflv_2332|M.gilvum_PYR-GCK RLSPLGERLVGPAREILRQVEDFKAAATQGVRGLA-PLRVSATA-YAPSD
Mvan_4315|M.vanbaalenii_PYR-1 RLTPLGQRLVGPAREILRQVEEFKAVATQAVRGLA-PVRVSATA-YAPSD
MSMEG_1902|M.smegmatis_MC2_155 RLTPLGEKLLGPARDILARVEDLKATASRITQGVA-PVRVGATA-YAPSD
Mb0121|M.bovis_AF2122/97 GLTREGERLVVWAKRILAEHAAFKAEVDAVRSGITGTLRLGTVP-TASTT
Rv0117|M.tuberculosis_H37Rv GLTREGERLVVWAKRILAEHAAFKAEVDAVRSGITGTLRLGTVP-TASTT
MLBr_02663|M.leprae_Br4923 GLTPEGERLVVWAKRILAEHDAFKAEVHAVRSGITGTLRLGTVP-TASTT
MMAR_0317|M.marinum_M GLTPEGERLVVWAKRIIAEHDAFKAEVDAVRSGITGTLRLGTVP-SASTT
MUL_4805|M.ulcerans_Agy99 GLTPEGERLVVWAKRIIAEHDAFKAEVDAVRSGITGTLRLGTVR-SASTT
MAB_3208c|M.abscessus_ATCC_199 RLTEFGKALLPLAGAALRTVGEIRSAADAMAGVLCGQLAIGTVP-HPSPL
MAV_3102|M.avium_104 QLTEEGERLRASTAAPLRQLELAMQYAGSPIARLARGVRLGILDTTVDVF
TH_3504|M.thermoresistible__bu SLTTAGRTLLAEGKPLLAAARTVAEHVRQTEAAAQAHYVIGHTPALSGAE
*: *. * : . :.
Mflv_2332|M.gilvum_PYR-GCK LLGELEAVLAGLPGPTEFSVPGSAAEVTAKLIAGQSELGLIHLPA-TDKR
Mvan_4315|M.vanbaalenii_PYR-1 LLGELEAAIASLPVPTEFSVPGSAAEVTAKLIAGQAELGLIHLPA-NDKR
MSMEG_1902|M.smegmatis_MC2_155 FLARLEKAVAGLAAPTQFSVPGSAAEVTAKLVSGHLDIGLIHLPS-ADKR
Mb0121|M.bovis_AF2122/97 ASLVLSAFCSAHPLAKVQVCSRLAATELYRRLREFELDAVIVHPE-TQDS
Rv0117|M.tuberculosis_H37Rv ASLVLSAFCSAHPLAKVQVCSRLAATELYRRLREFELDAVIVHPE-TQDS
MLBr_02663|M.leprae_Br4923 ASLVLSAFCSEHPLIKVQICSRLPVTELHRRLGDFELDAVIVHSA-PDDT
MMAR_0317|M.marinum_M ASLVLSDFCSAHPLVKAQICSRLAVTELYRRLRDFELDAAIVYTS-SDDG
MUL_4805|M.ulcerans_Agy99 ASLVLSDFCSAHPLVKAQICSRLAVTELYRRLRDFERDAAIVYTS-SDDG
MAB_3208c|M.abscessus_ATCC_199 LAEALAAYRRAHPAVTVTLTEARSDQLATAVVERRLDMAVMAWLAQPPSQ
MAV_3102|M.avium_104 AAHLLSSLAAAFPKVTFSVETGSTDQLVEAMLKGAVDVAVINPVP--DDR
TH_3504|M.thermoresistible__bu AYALIEPAVTAYPSTSFTLRQLYPDALTDAVFDGTVQLGLRRGVV-PTPE
: . . . . .
Mflv_2332|M.gilvum_PYR-GCK LRHRVVASYQGGVAVRFDDPLAAKDLVSIEELRDREVAIDFARPNPVVLA
Mvan_4315|M.vanbaalenii_PYR-1 LRYKVVASYQGGIAVRFDDPLAAKDLVSIEELRDREVAIDVARPNPVVLA
MSMEG_1902|M.smegmatis_MC2_155 LRHRVVARYQGAIAVRFDDPLAAKDVVAIEELRDREVVIDFARPNPVVLA
Mb0121|M.bovis_AF2122/97 DDVDLVPLYEEQYVLLSPADMLPPGTSTLVWRDAAQLPLALLTADMRDRQ
Rv0117|M.tuberculosis_H37Rv DDVDLVPLYEEQYVLLSPADMLPPGTSTLVWRDAAQLPLALLTADMRDRQ
MLBr_02663|M.leprae_Br4923 HAVDLVPLYHEHYVLVSPADMLPPGASVLAWPEASQLPLALLTPDMRDRQ
MMAR_0317|M.marinum_M HDVDLVPLYEERYVLLAPPEMLPPGVSTLSWLDAAQLPLALLSSDMRDRQ
MUL_4805|M.ulcerans_Agy99 HDVDLVPLYEERYVLLAPPEMLPPGVSTLSWLDAAQLPLALLSSDMRDRQ
MAB_3208c|M.abscessus_ATCC_199 LRERILAKENIVAVVHRDDPLAGRESVALAELRDRALISHPQGCEIR--T
MAV_3102|M.avium_104 IFYRDLLTEDLVVVGAATSGLDSGQMMPFTELVELPLVVPRSHTGIG--N
TH_3504|M.thermoresistible__bu LATAVIGYHRVRVAFVDSHPLAARESVDITELAREQVALWAPPGTSYYSD
: . : : :
Mflv_2332|M.gilvum_PYR-GCK GLTRRLNRKGVHHIVRTTNQRGGEVEMATQVFNRHLVAVVSYAPESFIGK
Mvan_4315|M.vanbaalenii_PYR-1 GLTRGLNRKGVHRIVRTTNQRGGEVEMATQVFNRHLVAVVSYAPDSFIGK
MSMEG_1902|M.smegmatis_MC2_155 GLTRGLNAKGVTKIVRAINQLGGELEMATQVFNRHLVAMVTYAPESMIGR
Mb0121|M.bovis_AF2122/97 VIDAAFADHAVSAIPQVETDSVASLFAQVATGNWASIVPHTWLWAMPMSG
Rv0117|M.tuberculosis_H37Rv VIDAAFADHAVSAIPQVETDSVASLFAQVATGNWASIVPHTWLWAMPMSG
MLBr_02663|M.leprae_Br4923 LIDRAFAGHAITATPQVETDSVASLFAQVATGNWACIVPHTWLWTTPIG-
MMAR_0317|M.marinum_M IIDAAFADHAITVSPQVETDSVASLFAQVATGNSASIVPHTWLWTTPAG-
MUL_4805|M.ulcerans_Agy99 IIDAAFADHAITVSPQVETDSVASLFAQVATGNSASIVPHTWLWTTPGR-
MAB_3208c|M.abscessus_ATCC_199 ALDQACLATGFT--PRVVFETSSADMMRHLVVRDLGVAVTPRLPREQLG-
MAV_3102|M.avium_104 VIENAALRRKVKISWRIATDSLALTKRLVAAGLVYGVLPLSACLNEIDSN
TH_3504|M.thermoresistible__bu FLMGACRRAGFEPDYVVSRVQGAATVAAPLTTGGIAFVTAAAGPTMNGR-
: . . . . .
Mflv_2332|M.gilvum_PYR-GCK IFSPPEFKLIPVDESTWAPAEIALAWVPERVQGR-------LAEVELMVE
Mvan_4315|M.vanbaalenii_PYR-1 IFSPPEFKLIPLDESTWAPAEIALAWVPERVQAR-------LTEVELMVE
MSMEG_1902|M.smegmatis_MC2_155 IFSPPEFKLIPIDEATWAPAQIALAWAPDRLSDRRSERVLRPAEIEALVD
Mb0121|M.bovis_AF2122/97 PTG-GEIRAVELVDPVLK-AQIALATNALGPGSPVARALITCAQALALNE
Rv0117|M.tuberculosis_H37Rv PTG-GEIRAVELVDPVLK-AQIALATNALGPGSPVARALITCAQALALNE
MLBr_02663|M.leprae_Br4923 ----AEIRAVEMVDPVLK-ADVALATNSAGPGSPVARALVASAKQLALNE
MMAR_0317|M.marinum_M ----GEIRPVQLVSPVVK-AQVALATSSTGPGSPVARALMTSAQGLALDE
MUL_4805|M.ulcerans_Agy99 --------------------------------------------------
MAB_3208c|M.abscessus_ATCC_199 -----RLRQIDLSDRAMS-GRLSLLWRGKG-------ATLEAVAFAACID
MAV_3102|M.avium_104 QLRYTPLTEPPLTHRVGVAATSNLELPREVTAKVGNILREETAHLIKTGR
TH_3504|M.thermoresistible__bu ------VQVVDLTPPLLVPVQALWQR----------------HTLSAIRD
Mflv_2332|M.gilvum_PYR-GCK QIADRLGPVHRGA---
Mvan_4315|M.vanbaalenii_PYR-1 QIADRLGPVHRGA---
MSMEG_1902|M.smegmatis_MC2_155 ELERAVARPAGGA---
Mb0121|M.bovis_AF2122/97 FFDTQLRGITRRR---
Rv0117|M.tuberculosis_H37Rv FFDTQLRGITRRR---
MLBr_02663|M.leprae_Br4923 FFEAQLAGITRRR---
MMAR_0317|M.marinum_M FFQTRLMEISRRR---
MUL_4805|M.ulcerans_Agy99 ----------------
MAB_3208c|M.abscessus_ATCC_199 ELCRKDANLVS-----
MAV_3102|M.avium_104 WPARLLSPQPWDPAVT
TH_3504|M.thermoresistible__bu RILEGLG---------