For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNSSTHTDVVIVGAGPNGLLLAGELALAGVRPVVLERLPQPSPELKANGIVGQVHRILDMRGLYQLLTGS SAPPQPLDGYIFAGMRVPFTGVDDNPMYGMFIKQPQVVRQLADWVRDLGVEIRWGHGLADMAPRPDGVTL NVTSPTGNHRLHATYLVGADGGRSTVRKAMGVGFPGVTSPVVARVAHVSIPDRLRGSGALEVPGFGRIPF GHNRFDHGVFMYMEFEPGRPLVGTLEYGALLPADAPAVTVDELRDSVTRILGVEVPIGPPSGPGPHALRR INGQNTRQAEQYRVGNVFLIGDAAHVHSAVGGPGLNLGMQDATNLGWKLAAAVHGWAPSGLLDTYHGERH PAGERVLMQSMAQTALLAGGPEVGALRTLFGELLATTDGAEHIAHVLAGADVRYDVGDGHPLSGRLVPEV VFDDGRRLADLMRAARPVLLDLDGGAYAAAAIDWRDRVDVITAAAAERPAPALLIRPDGYLAWAATGSGA DEHQRMRRALGRWFGEAHAGLAHA
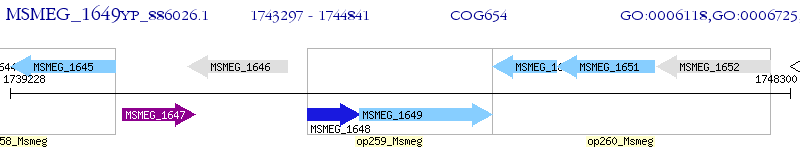
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1649 | - | - | 100% (514) | FAD dependent oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_2252 | - | 1e-73 | 36.93% (501) | flavin-type hydroxylase |
| M. smegmatis MC2 155 | MSMEG_4866 | mhpA | 1e-35 | 32.11% (383) | 3-(3-hydroxyphenyl)propionate hydroxylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_5009 | - | 1e-33 | 29.31% (539) | monooxygenase, FAD-binding |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0857 | - | 2e-76 | 37.14% (517) | hypothetical protein MAB_0857 |
| M. marinum M | MMAR_3439 | - | 1e-168 | 57.54% (504) | hydroxylase |
| M. avium 104 | MAV_2736 | - | 4e-76 | 38.88% (517) | hypothetical protein MAV_2736 |
| M. thermoresistible (build 8) | TH_1880 | - | 2e-77 | 37.85% (502) | flavin-type hydroxylase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2422 | - | 5e-76 | 37.24% (521) | hypothetical protein Mvan_2422 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1649|M.smegmatis_MC2_155 -------------------------------MNSSTHTDVVIVGAGPNGL
MMAR_3439|M.marinum_M --------------------------------MVDVAADVVISGAGPNGL
MAV_2736|M.avium_104 MRGVRFSE-----------------RSVRG-------HAVLVAGAGPTGL
Mvan_2422|M.vanbaalenii_PYR-1 MTTTRHKDTHKDTREDTREDTRDDSRDDGGDPNGDTDHAVVVGGGGPTGM
MAB_0857|M.abscessus_ATCC_1997 MAE----------------------------------HAVVVAGAGPTGL
TH_1880|M.thermoresistible__bu ------------------------------------VYDVIVVGGGPTGM
Mflv_5009|M.gilvum_PYR-GCK -------------------------------MPGAHRSDVVVVGAGPTGL
*:: *.**.*:
MSMEG_1649|M.smegmatis_MC2_155 LLAGELALAGVRPVVLERLPQPS-PELKANGIVGQVHRILDMRGLYQLLT
MMAR_3439|M.marinum_M MLACELALAGIRPVVLDKLSGPS-AEPKANGLVGQVVRMLDMRGLYQSFS
MAV_2736|M.avium_104 MLAGELGLAGVDVALVERRADQQLIGSRAGGLHARTLEILDQRGIVDRFL
Mvan_2422|M.vanbaalenii_PYR-1 MTAAELALAGVDVAVVERRTTPEITGSRAGGLLSRTLEILDQRGVVDRFL
MAB_0857|M.abscessus_ATCC_1997 MLAGELALAGIDVALLERRGSHEEVGSRAAGLHSRTIEVFDQRGIAERFL
TH_1880|M.thermoresistible__bu MLAGELRLHGVRVLVVEK-NSEPPAHVRSLGLHVRSIEMLDQRGLLDRFL
Mflv_5009|M.gilvum_PYR-GCK TLACCLRLHGVSVRVLDAAAGPA-TTSRANFLHARGSEVLDRMGALGTLP
* * * *: ::: :: : : .::* * :
MSMEG_1649|M.smegmatis_MC2_155 GSSAPPQPLDGYIFAGMRVPFTGVDD----NPMYGMFIKQPQVVRQLADW
MMAR_3439|M.marinum_M GDDGPPQPAQGWMFAAMPLSFLGLND----NSLYTMLMPQPRLVRLLEQR
MAV_2736|M.avium_104 AEGQVAQVAG---FAQIRLDISDFPT----RHPYGLALWQNHIERILADW
Mvan_2422|M.vanbaalenii_PYR-1 SAGETMQVHG---FAQVRLDVSDLPT----RHPYGLALWQKHIERILAEW
MAB_0857|M.abscessus_ATCC_1997 AQGRTGQVTH---FSGIFMSIEDFPT----RHPYALGLLQHYTEKTLAAW
TH_1880|M.thermoresistible__bu AHGQRHPVGGF--FAAIAKPPAELDT----AHGYVLGIRQPVTDRLLAEW
Mflv_5009|M.gilvum_PYR-GCK QESLRALRITN-YLGDRPLATLEFGDPGMRTAAPPMVVSQAKVEAALRAR
. :. . : : * *
MSMEG_1649|M.smegmatis_MC2_155 VRDLGVEIRWGHGLADMAPRPDGVTLNVTSPTGNHRLHATYLVGADGGRS
MMAR_3439|M.marinum_M ASALGVDLRWGDELTGFRCDSQSVVAEVSSPQRSYEISSRYLIGADGGRS
MAV_2736|M.avium_104 VGELGVPIYRGHRIATCTQDDTGVDVVASD---GRTLRGQYLVGCDGGRS
Mvan_2422|M.vanbaalenii_PYR-1 VDELAVPVYRGREVVGVRQDGAGVDIELSD---GRVLRAGYLVGCDGGRS
MAB_0857|M.abscessus_ATCC_1997 IDELGVPIYYNAEAVGLGQDDVGVDVALAD---GSTLRGQYLVGCDGGRS
TH_1880|M.thermoresistible__bu AAERGAVIRRGCAVVGLDQDAAGITVELAD---GSVQRARYLVGCDGGRS
Mflv_5009|M.gilvum_PYR-GCK LADLDVEPEWGRKVIDVREDADGVVADLAD---GARVRGQWLVGCDGTTS
. . .: :. . . :*:*.** *
MSMEG_1649|M.smegmatis_MC2_155 TVRKAMGVGFPGVTSPVVARVAHVSIPDRLRGSG-ALEVPGFGRIPFGHN
MMAR_3439|M.marinum_M LVRKTLGIEFPGCTSATVGRLAHVQLPDEFRTADRAVEIPEFGRLPFGHS
MAV_2736|M.avium_104 VIRKAAGIDFPGWEATTSYLLAEAEMDWRTGQPPQWGIHHDSLGVHALSE
Mvan_2422|M.vanbaalenii_PYR-1 LIRKSAGIEFPGWEPTTSYLLAEVEIAPEDGREPEWGIRRDALGVHALSR
MAB_0857|M.abscessus_ATCC_1997 TIRKAAGIGFPGHDPSCSAMLVDAEIT---EQPLEFGLLRKPGQAFLGIL
TH_1880|M.thermoresistible__bu TVRKLAGIGFPGEPARSETLLGEMSLSADPATVAAVVAEVRKTQLRFGAM
Mflv_5009|M.gilvum_PYR-GCK VVRRQTGIEFPGVRLSERFLLADIHLDWAVDRAGTTGWIHPDGVVGAMPM
:*: *: *** : . :
MSMEG_1649|M.smegmatis_MC2_155 RFDHGVFMYMEFEPGRPLVGTLEYGALLPADAPAVTVDELRDSVTRILGV
MMAR_3439|M.marinum_M RFDRGGVIFAEFEPGRSMLGTIEFGP--PVEERPMSLAELRESVQRVLGV
MAV_2736|M.avium_104 TEPAGGVRVLVTERQLR-------------SAAEPNLDDLRQALIAVYRT
Mvan_2422|M.vanbaalenii_PYR-1 SD-DGPVRIMVTEQDLR-------------RSGVPDLAELREALVTVYGT
MAB_0857|M.abscessus_ATCC_1997 PFEEGWCRLAFSTGLDR-------------LGVEPTLEEAKEALIAIAGT
TH_1880|M.thermoresistible__bu PLGDGVYRVIVPAEGVAG------------DRAEPTLDEFRRRLKAFAGT
Mflv_5009|M.gilvum_PYR-GCK PSTDGRDDLWRLFVYDPS------------LDRKPTDSEILDRISELVPY
* : : .
MSMEG_1649|M.smegmatis_MC2_155 EVPIGPPSGPGPHALRRINGQNTRQAEQYRVGNVFLIGDAAHVHSAVGGP
MMAR_3439|M.marinum_M DIPLKRPKGPGPHALRRINGQNTRHAERYRRGRVLLLGDAAHVHSPMGGP
MAV_2736|M.avium_104 DYGIHSPR-----WISRFT-DAARQAARYRDRRVLLAGDAAHIHHPVGGQ
Mvan_2422|M.vanbaalenii_PYR-1 DFGIHSPT-----SISRFT-DTARQAVAYRSGRILLVGDAAHVHHPVGGQ
MAB_0857|M.abscessus_ATCC_1997 DFGIHNPR-----WISRFS-DMTRQADRYRDGRVFLAGDSAHIHFPFGGQ
TH_1880|M.thermoresistible__bu DFGAHTPR-----WLSRFG-DATRLAERYRAGRVFLAGDAAHVHPPAGGQ
Mflv_5009|M.gilvum_PYR-GCK RTGRRVKIG-DAEWLSMFT-VHRRLADTYRRGRVLIAGDAAHVHAPFGGQ
: : * * ** .::: **:**:* . **
MSMEG_1649|M.smegmatis_MC2_155 GLNLGMQDATNLGWKLAAAVHGWAPSGLLDTYHGERHPAGERVLMQSMAQ
MMAR_3439|M.marinum_M GLNLGLQDTFNLGWKLAAELNGWAPDGLLDTYESERHPVGARVMMHSLAQ
MAV_2736|M.avium_104 GLNTGVQDAVNLGWKLALVVGGASPPALLDTYHAERHPVGERVLRNAIAQ
Mvan_2422|M.vanbaalenii_PYR-1 GLNTGMQDAVNLGWKLAQVVQGWSPDTLLDTYHDERQPVGARVLRNAVAQ
MAB_0857|M.abscessus_ATCC_1997 GLNTGIQDAVNLGWKLAAVIKGHAPDALLDTYHLERHPVAERVLHNTMAQ
TH_1880|M.thermoresistible__bu GLNLGIQDAFNLGWKLAAAVGGWAPDGLLDTYHAERHPVAADVLNNTRAQ
Mflv_5009|M.gilvum_PYR-GCK GMLTGIGDAENVGFKLALVVRGLAADDLLDTYEAERRPLATQVLRGTSAI
*: *: *: *:*:*** : * :. *****. **:* . *: : *
MSMEG_1649|M.smegmatis_MC2_155 TALLAGG-PEVGALRTLFG-ELLATTDGAEHIAHVLAGADVRYD-----V
MMAR_3439|M.marinum_M TALMAPG-PEVGALRELFG-ELLTIPEVTKQMGALLAGSDLIYP-----V
MAV_2736|M.avium_104 MALLRPD-ERTRALRDLVT-ELLGMDEPRKRFAAMISGLDVRYD-----L
Mvan_2422|M.vanbaalenii_PYR-1 MALLRPD-DRTTVLHGVVS-DMLDMDEPRRRFAAMMSGLDIHYD-----L
MAB_0857|M.abscessus_ATCC_1997 TPLSEAG-PRIDALRDIVA-DLLAMDEPRKRIAGMMSGLDIHYM-----L
TH_1880|M.thermoresistible__bu MELMSPE-PGPRAVRWLLA-ELMDFEEVNRYLTEKITAIGVRYHT----M
Mflv_5009|M.gilvum_PYR-GCK TRVNVAGNPIVRFLRDRVAPRVFGLPAVQRWTTYTASQLWVSYRNGPLGG
: :: . :: . : : *
MSMEG_1649|M.smegmatis_MC2_155 GDGHPLSGRLVPEVVFDDGR---RLADLMRAARPVLLDLDGGAYAAAAID
MMAR_3439|M.marinum_M GDEHPLSGRLVPVLTTDDGR---RIDELMCGARPLLIDLADGDAAAAAQG
MAV_2736|M.avium_104 GPGHPLLGRRMPDLDLVTADGASRVFALLRNARPVLLNFAGAGRRSVAG-
Mvan_2422|M.vanbaalenii_PYR-1 GDGHPLLGLRMPDLDLETAHGPTRVSALLRQARPVLLDLGAPDAVDLTG-
MAB_0857|M.abscessus_ATCC_1997 GDGHQLLGRRMPDLDVVTADGAVRVYEHLHTARPVLFNFGARQILNVEG-
TH_1880|M.thermoresistible__bu GDGHGLLGRRMRDIPLTGG----RLYARMHAGRGVLLDRTG--TLSVSG-
Mflv_5009|M.gilvum_PYR-GCK RGSKPRPGDRIDDAPCSRPDGTATRLHGELGGRWALLLPRGVPAVGAAAV
: * : .* *:
MSMEG_1649|M.smegmatis_MC2_155 WRDRVDVITAA----------AAERPAPALLIRPDGYLAWAATGSGADEH
MMAR_3439|M.marinum_M WADRVEIVHTS----------IADRPAAAMLIRPDGYVAWAANAFDTGAR
MAV_2736|M.avium_104 WADRVRVVDAKCVADWELPVLGRVSAPPAVLIRPDGHVAWAGDPN----D
Mvan_2422|M.vanbaalenii_PYR-1 WEARVRMVRANAATVWELPVIGEVPAPAAVLIRPDGHVAWVAGDS----A
MAB_0857|M.abscessus_ATCC_1997 WEDRVHVVDAKYSGIWELPVLGEVSAPDAVLVRPDGYVAWVGEGS----A
TH_1880|M.thermoresistible__bu WADRVDHIVDP----------GAALDVPAVLLRPDGHVAWVGADHPG--Q
Mflv_5009|M.gilvum_PYR-GCK RGPAGYLADHVVT--------LHHDRDDVMLVRPDAHLAWRGAHDDP---
.:*:***.::** .
MSMEG_1649|M.smegmatis_MC2_155 QRMRRALGRWFGEAHAGLAHA
MMAR_3439|M.marinum_M GSLRDALQRWFGSHLGG----
MAV_2736|M.avium_104 PGLHDALTTWCGPPAAGG---
Mvan_2422|M.vanbaalenii_PYR-1 AAPAEAAARWFGAPTGRR---
MAB_0857|M.abscessus_ATCC_1997 DGLRGALTRWVGSPSRA----
TH_1880|M.thermoresistible__bu QDLTARLSRWFGAPA------
Mflv_5009|M.gilvum_PYR-GCK ----TGLDRWLATALHPGGTQ
* .