For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSEPLLLQDRDERGVVTLTLNRPQAFNALSEAMLAALGEAFGTLAEDESVRAVVLAASGKAFCAGHDLKE MRAEPSREYYEKLFARCTDVMLAIQRLPAPVIARVHGIATAAGCQLVAMCDLAVATRDARFAVSGINVGL FCSTPGVALSRNVGRKAAFEMLVTGEFVSADDAKGLGLVNRVVAPKALDDEIEAMVSKIVAKPRAAVAMG KALFYRQIETDIESAYADAGTTMACNMMDPSALEGVSAFLEKRRPEWHTPQPSTA
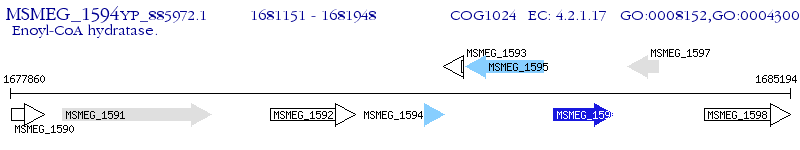
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1594 | - | - | 100% (265) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_6774 | - | 1e-28 | 33.60% (247) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_3139 | - | 1e-28 | 32.20% (264) | enoyl-CoA hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1099c | echA8 | 2e-29 | 31.40% (258) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_2444 | - | 3e-33 | 36.58% (257) | enoyl-CoA hydratase/isomerase |
| M. tuberculosis H37Rv | Rv1070c | echA8 | 2e-29 | 31.40% (258) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 2e-30 | 32.17% (258) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_4588c | - | 1e-29 | 34.10% (261) | enoyl-CoA hydratase/isomerase |
| M. marinum M | MMAR_4396 | echA8 | 5e-31 | 32.56% (258) | enoyl-CoA hydratase EchA8 |
| M. avium 104 | MAV_2641 | - | 3e-32 | 36.25% (251) | enoyl CoA hydratase |
| M. thermoresistible (build 8) | TH_4102 | - | 7e-29 | 30.86% (256) | PUTATIVE short chain enoyl-CoA hydratase |
| M. ulcerans Agy99 | MUL_0210 | echA8 | 3e-31 | 32.56% (258) | enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_4207 | - | 9e-33 | 36.61% (254) | enoyl-CoA hydratase/isomerase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4396|M.marinum_M ---MSYETILVERDQRVGIITLNRPQALNALNSQVMNEVTSAAAELDNDP
MUL_0210|M.ulcerans_Agy99 ---MSYETILVERDQRVGIITLNRPQALNALNSQVMNEVTSAAAELDNDP
Mb1099c|M.bovis_AF2122/97 ---MTYETILVERDQRVGIITLNRPQALNALNSQVMNEVTSAATELDDDP
Rv1070c|M.tuberculosis_H37Rv ---MTYETILVERDQRVGIITLNRPQALNALNSQVMNEVTSAATELDDDP
MLBr_02402|M.leprae_Br4923 ---MKYDTILVDGYQRVGIITLNRPQALNALNSQMMNEITNAAKELDIDP
TH_4102|M.thermoresistible__bu MSAKNYETILVTREDRVGIITLNRPKALNALNSQVMTEVTDAAAEFDSDP
MAB_4588c|M.abscessus_ATCC_199 ----MADEVLIEQRDRVLLITINRPDARNAVNRAVSQGLAAAADQLDSSA
Mflv_2444|M.gilvum_PYR-GCK --MADTEVLLSSDRDGVRTLTLNRPHRRNALDARLWVELADALRALKREQ
Mvan_4207|M.vanbaalenii_PYR-1 --MVDDQVLLSTDRDGVRTLTLNRPERRNALDARLWVELADALRDLKRDH
MAV_2641|M.avium_104 --MNTQAAVLSDIADGVLTITLNRPEAANAVRPDDRDALIALLAAADADE
MSMEG_1594|M.smegmatis_MC2_155 --MSEPLLLQDRDERGVVTLTLNRPQAFNALSEAMLAALGEAFGTLAEDE
: * :*:***. **: : .
MMAR_4396|M.marinum_M GIGAIIITGSAKAFAAGADIK---------EMADLTFSEAFDADFFAAWG
MUL_0210|M.ulcerans_Agy99 GIGAIIITGSAKAFAAGADIK---------EMADLTFSEAFDADFFAAWG
Mb1099c|M.bovis_AF2122/97 DIGAIIITGSAKAFAAGADIK---------EMADLTFADAFTADFFATWG
Rv1070c|M.tuberculosis_H37Rv DIGAIIITGSAKAFAAGADIK---------EMADLTFADAFTADFFATWG
MLBr_02402|M.leprae_Br4923 DVGAILITGSPKVFAAGADIK---------EMASLTFTDAFDADFFSAWG
TH_4102|M.thermoresistible__bu GIGAIIVTGNEKAFAAGADIK---------EMADLEFADVFAADFFAPWS
MAB_4588c|M.abscessus_ATCC_199 DLSVAIITGAGGNFCAGMDLK---------AFVS---GEAVLSERGLGFT
Mflv_2444|M.gilvum_PYR-GCK DVRALVLTGAGGAFCSGADIG-------TGEQ--IHP-RYKLDRLTDVAL
Mvan_4207|M.vanbaalenii_PYR-1 DVRALVLTGAGGAFCSGADIG-------TGEQ--IHP-RYKLDRLTDVAL
MAV_2641|M.avium_104 KVRVVVLRANGKHFCSGADVGSLAERRARGEKRVLEPTRRIMNGAQKLVA
MSMEG_1594|M.smegmatis_MC2_155 SVRAVVLAASGKAFCAGHDLK------EMRAEPSREYYEKLFARCTDVML
: . :: . *.:* *:
MMAR_4396|M.marinum_M KLAAVRTPTIAAVAGYALGGGCEVAMMCDLIIAADTAKFGQPEIKLGVLP
MUL_0210|M.ulcerans_Agy99 KLAAVRTPTIAAVAGYALGGGCEVAMMCDLIIAADTAKFGQPEIKLGVLP
Mb1099c|M.bovis_AF2122/97 KLAAVRTPTIAAVAGYALGGGCELAMMCDVLIAADTAKFGQPEIKLGVLP
Rv1070c|M.tuberculosis_H37Rv KLAAVRTPTIAAVAGYALGGGCELAMMCDVLIAADTAKFGQPEIKLGVLP
MLBr_02402|M.leprae_Br4923 KLAAVRTPMIAAVAGYALGGGCELAMMCDLLIAADTAKFGQPEIKLGVLP
TH_4102|M.thermoresistible__bu KFAATRTPTIAAVAGYALGGGCELAMMCDLLIAADTAKFGQPEIKLGVLP
MAB_4588c|M.abscessus_ATCC_199 NVPP-RKPIIAAVEGFALAGGTELVLSCDLVVAGRSAKFGIPEVKRGLVA
Mflv_2444|M.gilvum_PYR-GCK ALHELAIPTIAKVTGIAVGAGWNLALGCDLVVATPESRFCQIFSKRGLSV
Mvan_4207|M.vanbaalenii_PYR-1 ALHELAVPTIAKVTGIAVGAGWNLALGCDLVVATPESRFCQIFSKRGLSV
MAV_2641|M.avium_104 SVLDCNKPVIAAVHGAATGLGAHVVYASDLVIATENAYFAESFVKRGLVV
MSMEG_1594|M.smegmatis_MC2_155 AIQRLPAPVIARVHGIATAAGCQLVAMCDLAVATRDARFAVSGINVGLFC
. * ** * * * . * .:. .*: :* : * : *:
MMAR_4396|M.marinum_M GMGGSQRLTRAIGKAKAMDLILTGRTIDAAEAERSGLVSRVVPADDLLTE
MUL_0210|M.ulcerans_Agy99 GMGGSQRLTRAIGKAKAMDLILTGRTIDAAEAERSGLVSRVVPADDLLTE
Mb1099c|M.bovis_AF2122/97 GMGGSQRLTRAIGKAKAMDLILTGRTMDAAEAERSGLVSRVVPADDLLTE
Rv1070c|M.tuberculosis_H37Rv GMGGSQRLTRAIGKAKAMDLILTGRTMDAAEAERSGLVSRVVPADDLLTE
MLBr_02402|M.leprae_Br4923 GMGGSQRLTRAIGKAKAMDLILTGRTIDAAEAERSGLVSRVVLADDLLPE
TH_4102|M.thermoresistible__bu GMGGSQRLTRAIGKAKAMDLILTGRTIDAEEAERSGLVSRVVPADKLLDE
MAB_4588c|M.abscessus_ATCC_199 GAGGLLRLPNRIPYQVAMELALTGESFTAEDAAKYGFINRLVDDGQALDT
Mflv_2444|M.gilvum_PYR-GCK DLGGSWLLPKTVGLQQAKRLVLLADMIDADEAHGLGLVTWVKPAGEIDGF
Mvan_4207|M.vanbaalenii_PYR-1 DLGGSWLLPRIVGLQQAKRLVLLAEMIGADEARDLGLVTWVKPSGEIDAF
MAV_2641|M.avium_104 DGGGCYLLPRRIGMQKAKEMAFFGEKLSATEAFTLGLVNRVVSAADFDAT
MSMEG_1594|M.smegmatis_MC2_155 STPG-VALSRNVGRKAAFEMLVTGEFVSADDAKGLGLVNRVVAPKALDDE
. * *.. : * : . . . * :* *::. :
MMAR_4396|M.marinum_M AKAVATTISQMSRSAARMAKEAVNRTFEATLAEGLLYERRLIHSAFATAD
MUL_0210|M.ulcerans_Agy99 AKAVATTISQMSRSAARMAKEAVNRTFEATLAEGLLYERRLIHSAFATAD
Mb1099c|M.bovis_AF2122/97 ARATATTISQMSASAARMAKEAVNRAFESSLSEGLLYERRLFHSAFATED
Rv1070c|M.tuberculosis_H37Rv ARATATTISQMSASAARMAKEAVNRAFESSLSEGLLYERRLFHSAFATED
MLBr_02402|M.leprae_Br4923 AKAVATTISQMSRSATRMAKEAVNRSFESTLAEGLLHERRLFHSTFVTDD
TH_4102|M.thermoresistible__bu AKETAATIAAMSLSAARMAKEAVNRAFETSLAEGLLFERRVFHSAFATAD
MAB_4588c|M.abscessus_ATCC_199 ALELAAKITANGPLAVAATKRIIIESASWAPEEAFAKQGEILMPIFVSED
Mflv_2444|M.gilvum_PYR-GCK IDELADRLAAGPPFALAQSKALLNDGATATLREALANEARAQPGNFATAD
Mvan_4207|M.vanbaalenii_PYR-1 VDDLADRLAGGPPFALAQSKALLNDGANATLREALANEARAQPGNFATAD
MAV_2641|M.avium_104 VADFAGRLAGAPTSAIAFTKRLLNDSPDTDRSGAFVAEAMAQEIQSYSHD
MSMEG_1594|M.smegmatis_MC2_155 IEAMVSKIVAKPRAAVAMGKALFYRQIETDIESAYADAGTTMACNMMDPS
. : * * . . .
MMAR_4396|M.marinum_M QSEGMAAFIEKRPPNFTHR-------------
MUL_0210|M.ulcerans_Agy99 QSEGMAAFIEKRPPNFTHR-------------
Mb1099c|M.bovis_AF2122/97 QSEGMAAFIEKRAPQFTHR-------------
Rv1070c|M.tuberculosis_H37Rv QSEGMAAFIEKRAPQFTHR-------------
MLBr_02402|M.leprae_Br4923 QSEGMAAFIEKRAPQFTHR-------------
TH_4102|M.thermoresistible__bu QSEGMAAFTEKRPPKFTHR-------------
MAB_4588c|M.abscessus_ATCC_199 AKEGAKAFAEKRAPVWQGK-------------
Mflv_2444|M.gilvum_PYR-GCK SAEAYAAFAAKRDPEFSGAWALYEQPEEQR--
Mvan_4207|M.vanbaalenii_PYR-1 SVEAYAAFAAKREPEFTGAWVQYEKPTQKDEE
MAV_2641|M.avium_104 STEGVQAFVEKRPTEFTG-W------------
MSMEG_1594|M.smegmatis_MC2_155 ALEGVSAFLEKRRPEWHTPQPSTA--------
*. ** ** . :